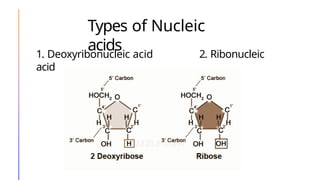
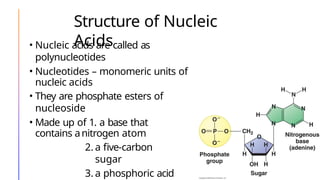
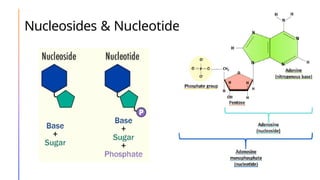
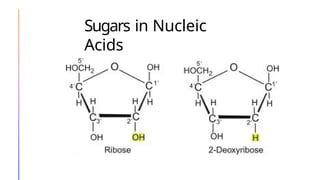
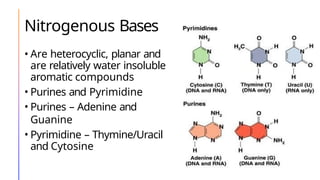
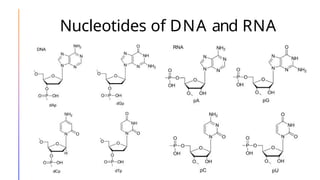
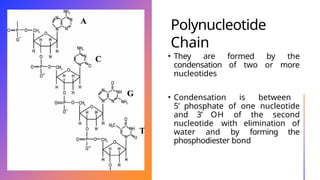
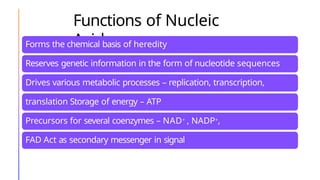
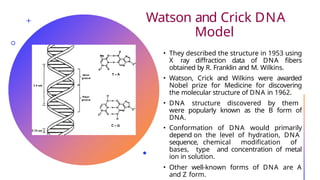
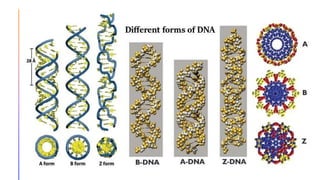
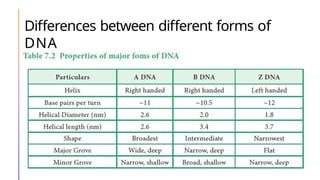
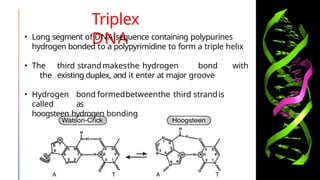
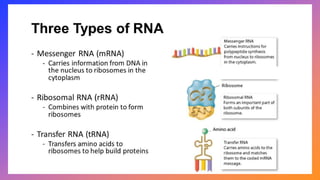
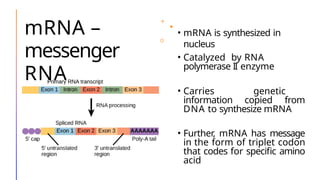
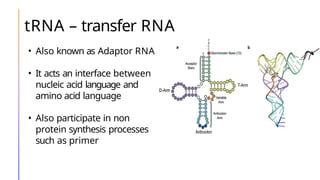
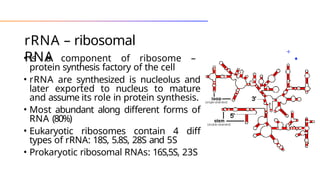
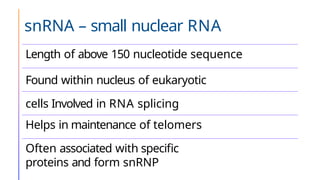
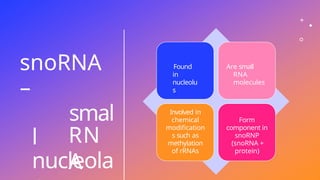
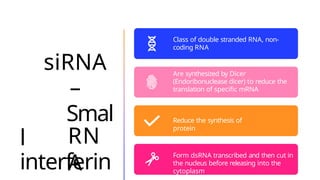
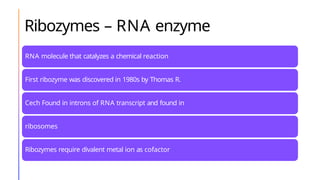
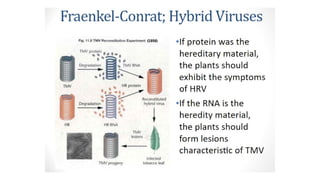
The document discusses nucleic acids, detailing their discovery, structure, and functions, including DNA and RNA types. It covers the history of nucleic acid research, key figures involved, and essential concepts like base pairing and the structure of DNA. It also delves into various RNA types and their roles in protein synthesis and gene expression regulation.