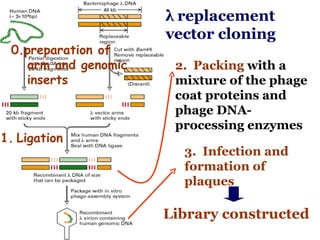
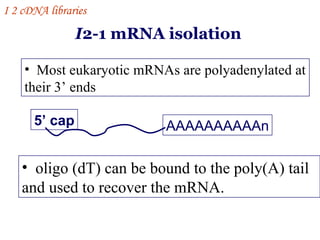
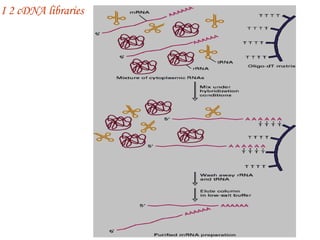
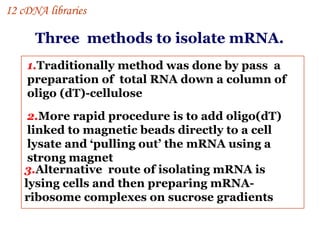
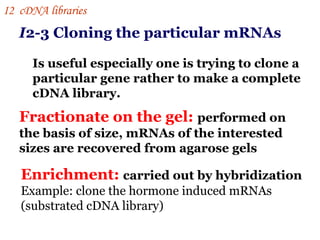
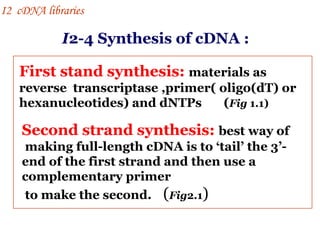
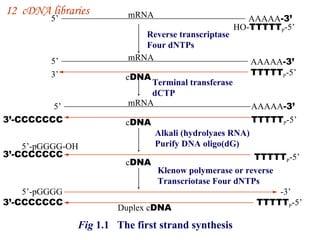
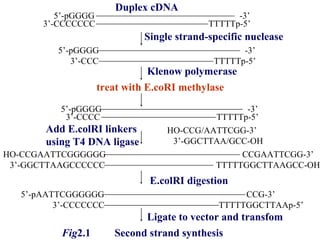
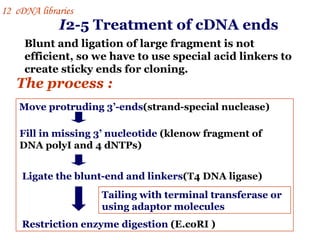
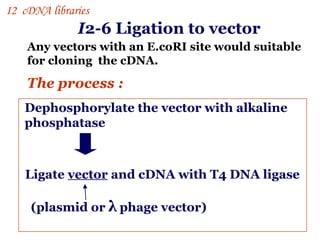
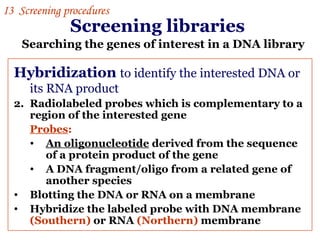
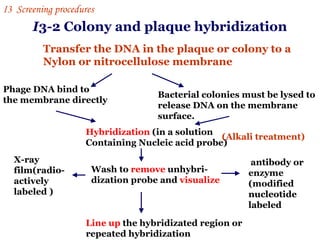
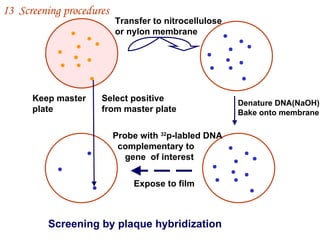
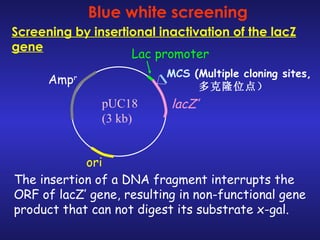
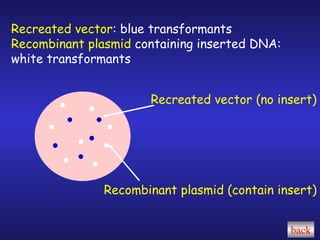
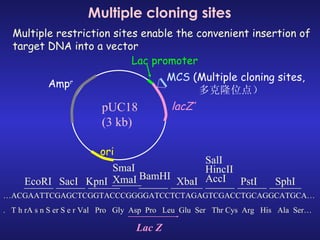
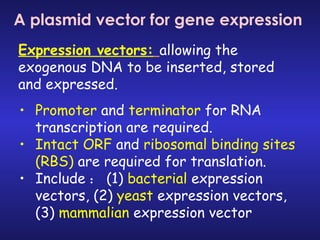
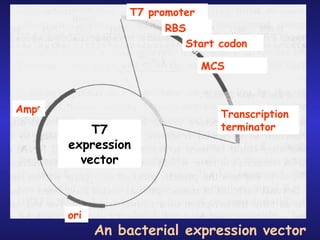
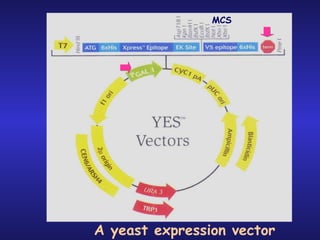
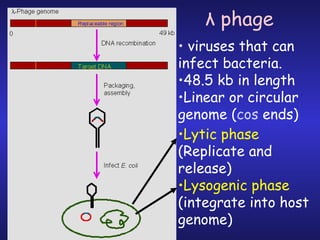
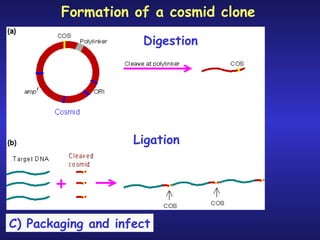
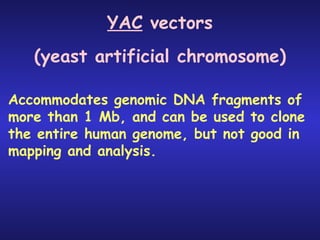
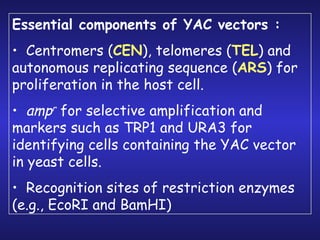
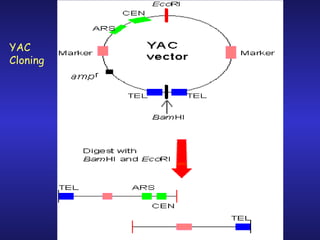
The document discusses gene cloning, expression, and functional study. It describes different types of vectors used for cloning genes, including cloning vectors, expression vectors, and integration vectors. It provides details on various cloning vectors such as plasmid vectors, bacteriophage vectors, cosmids, BACs, and eukaryotic vectors. It also describes expression vectors and components required for gene expression. Finally, it discusses bacteriophage vectors, cosmids, YAC vectors, and BAC vectors which are used to clone large DNA fragments from eukaryotes.





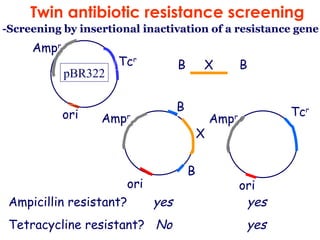
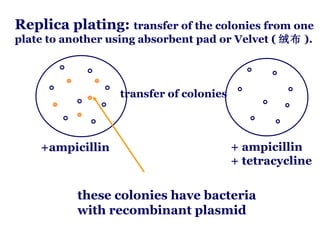






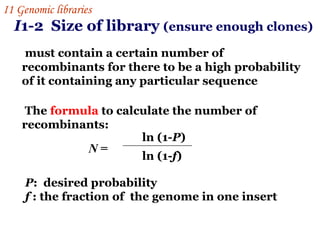
![For example :for a probability of 0.99 with insert sizes of 20 kb these values for the E.coli (4.6 ×10 6 bp) and human (3×10 9 bp) genomes are : N E.coli = = 1.1 ×10 3 ln( 1-0.99) ln[1-(2×10 4 /4.6×10 6 )] N human = = 6.9 ×10 5 ln(1-0.99) ln[1-(2 ×10 4 /3 ×10 9 )] These values explain why it is possible to make good genomic libraries from prokaryotes in plasmids where the insert size is 5-10kb ,as only a few thousand recombinants will be needed. I 1 Genomic libraries](https://image.slidesharecdn.com/application1-110516074210-phpapp01/85/Application1-27-320.jpg)