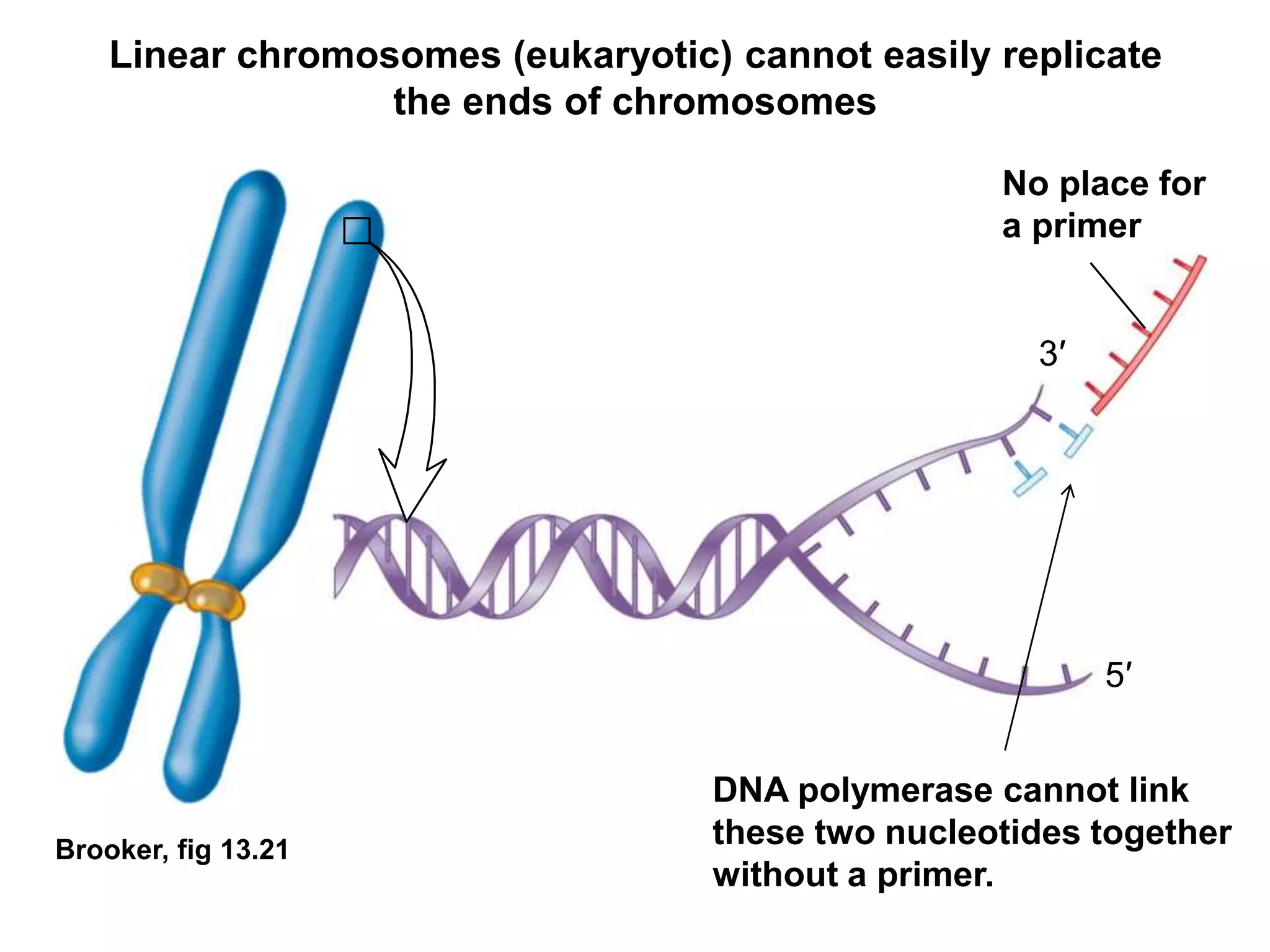
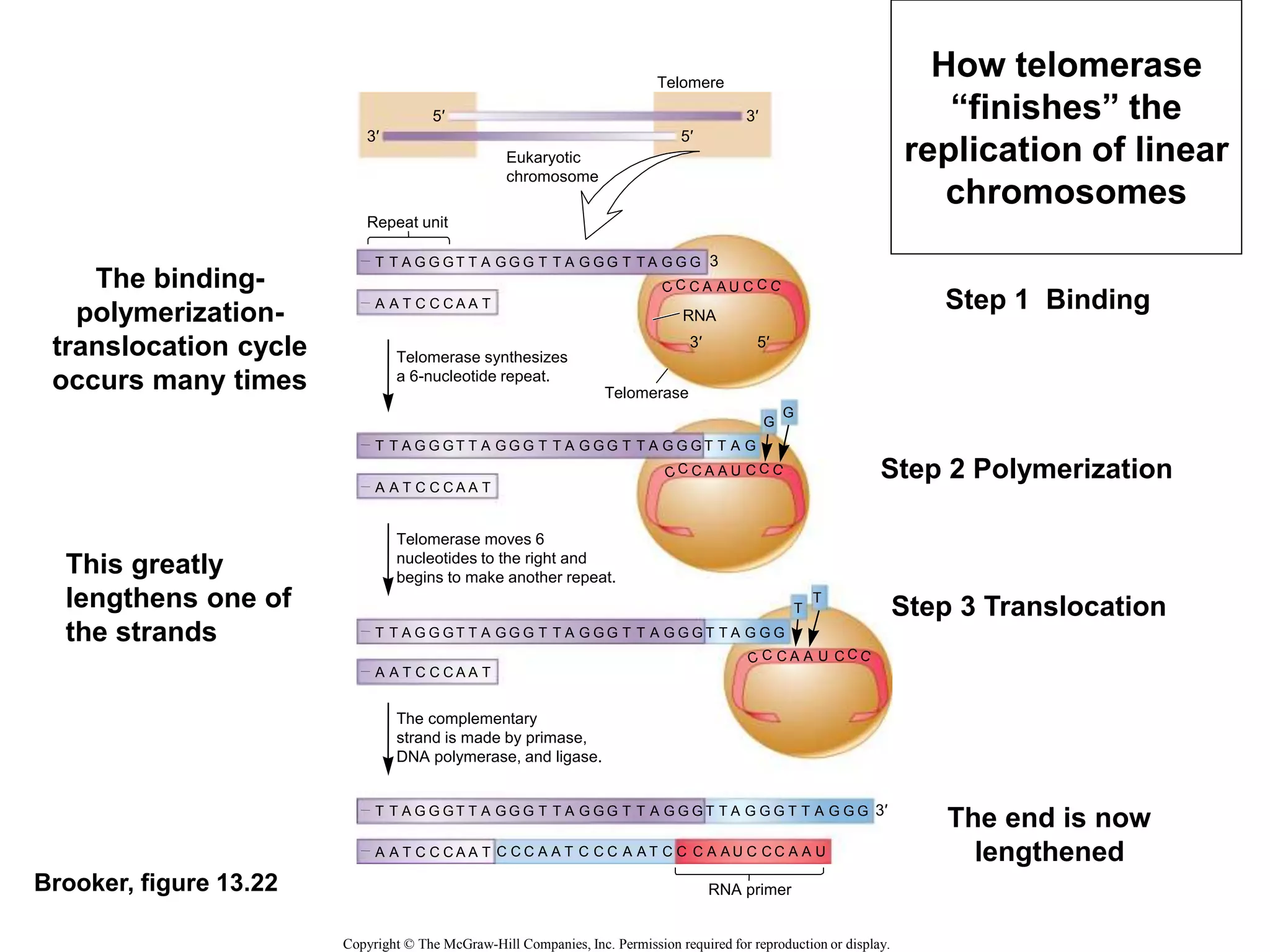
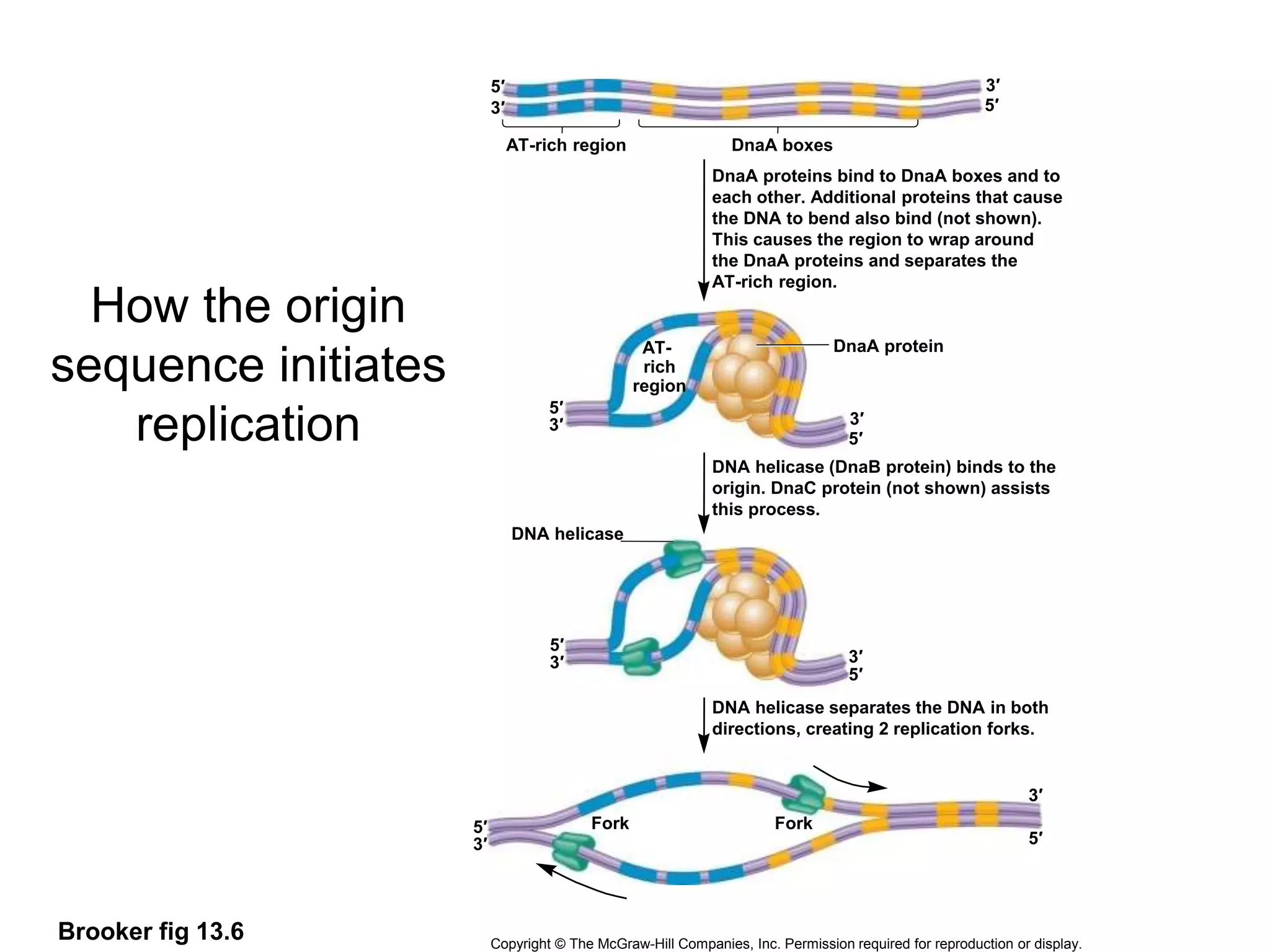
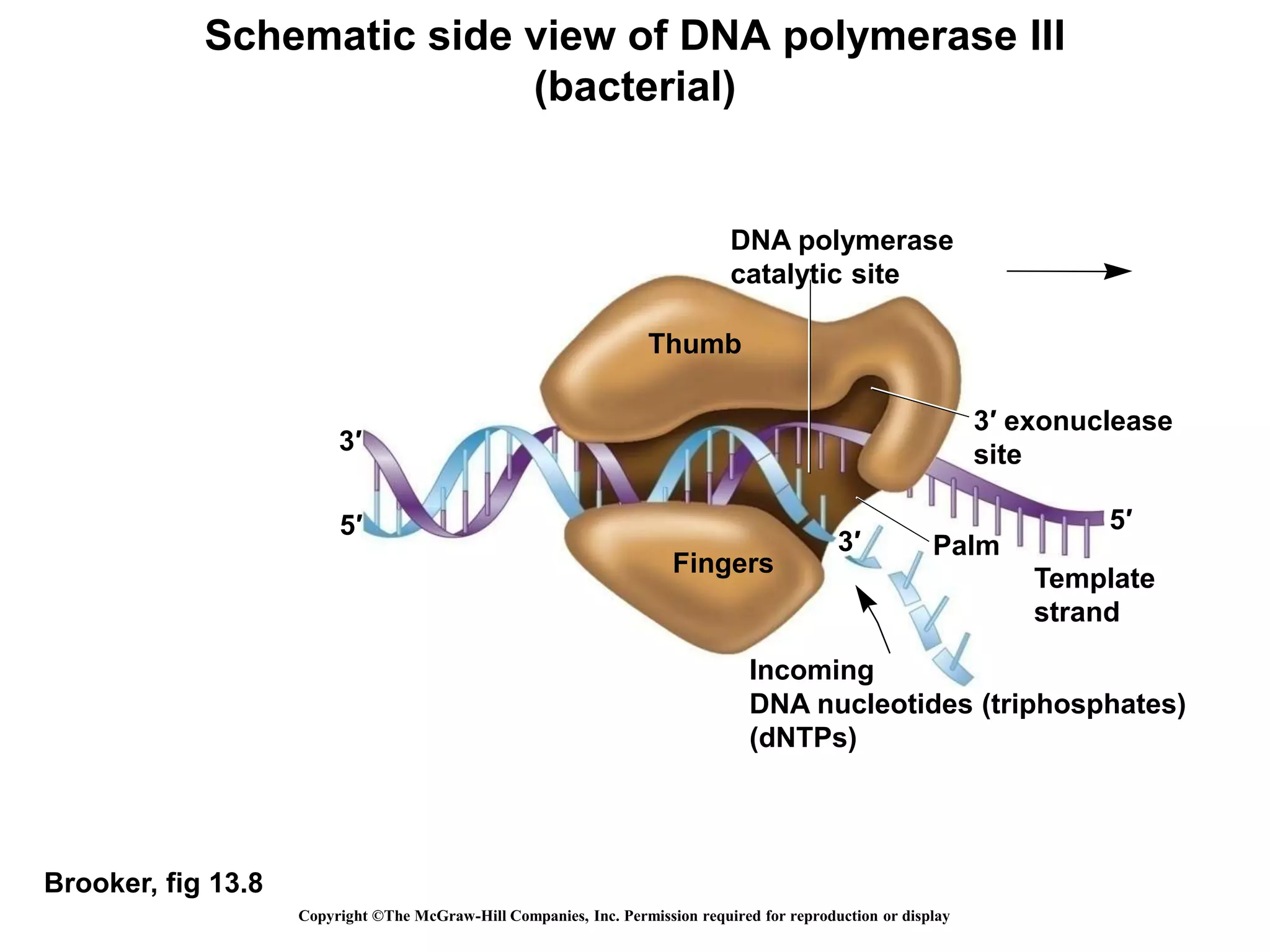
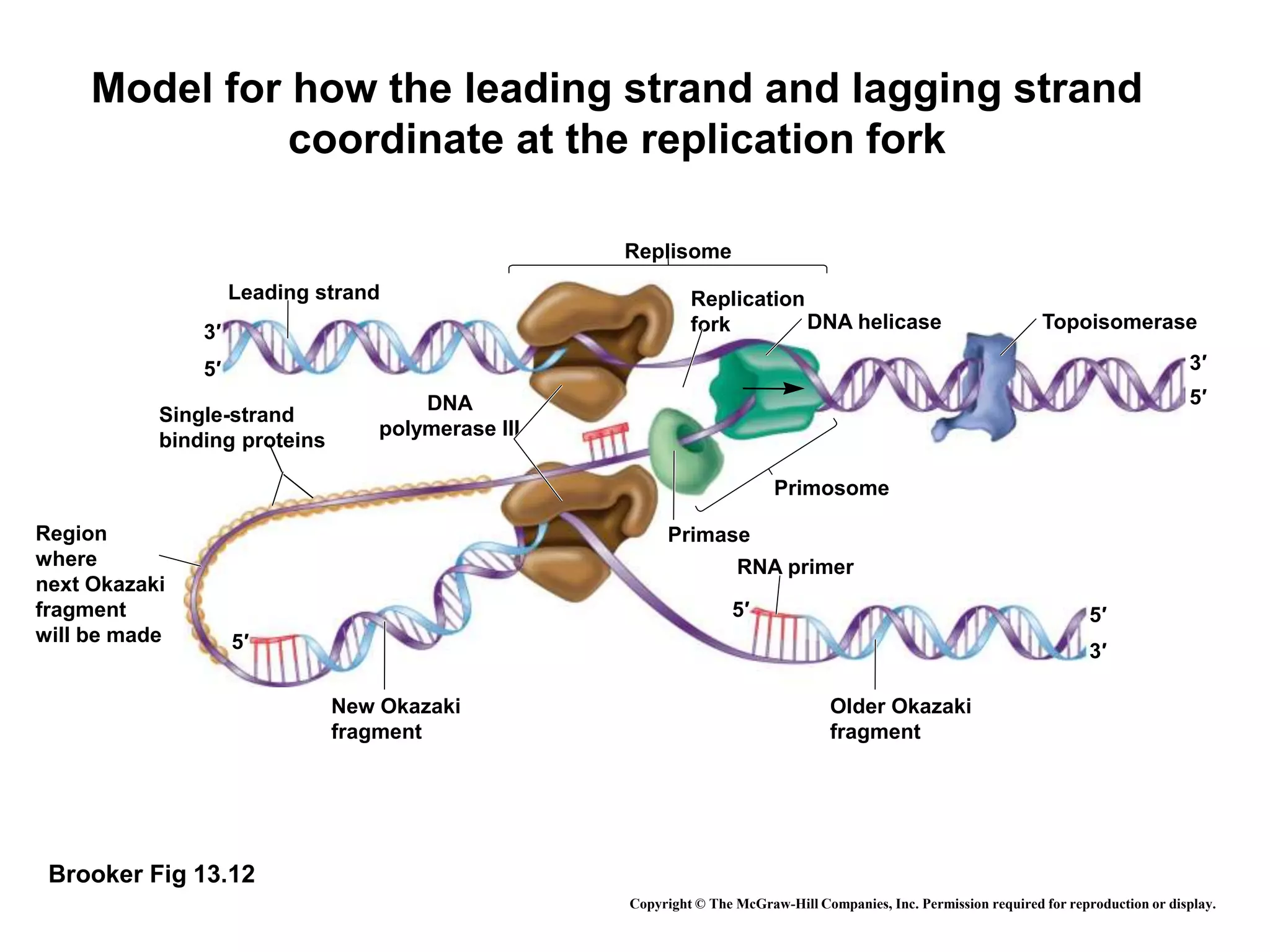
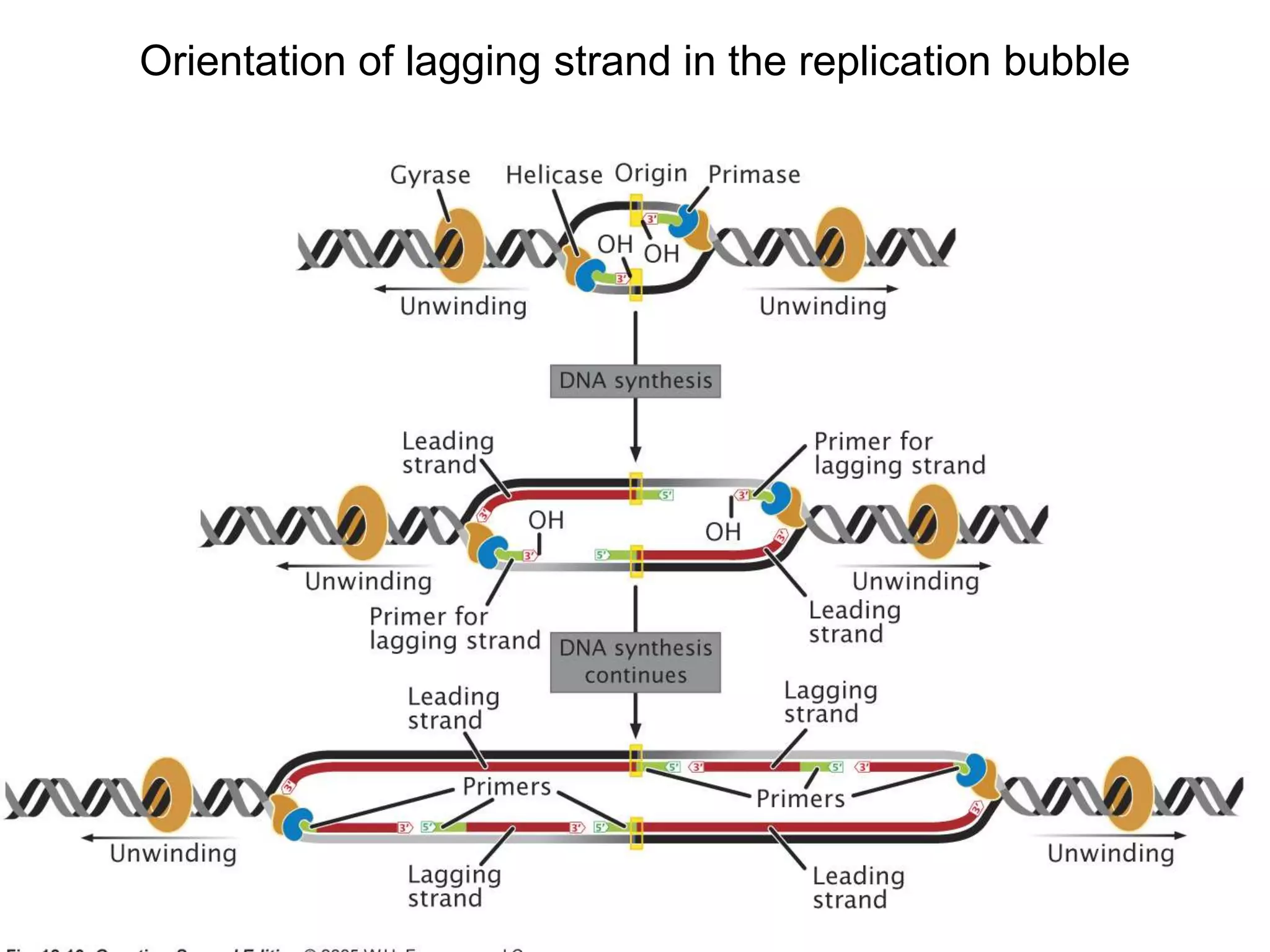
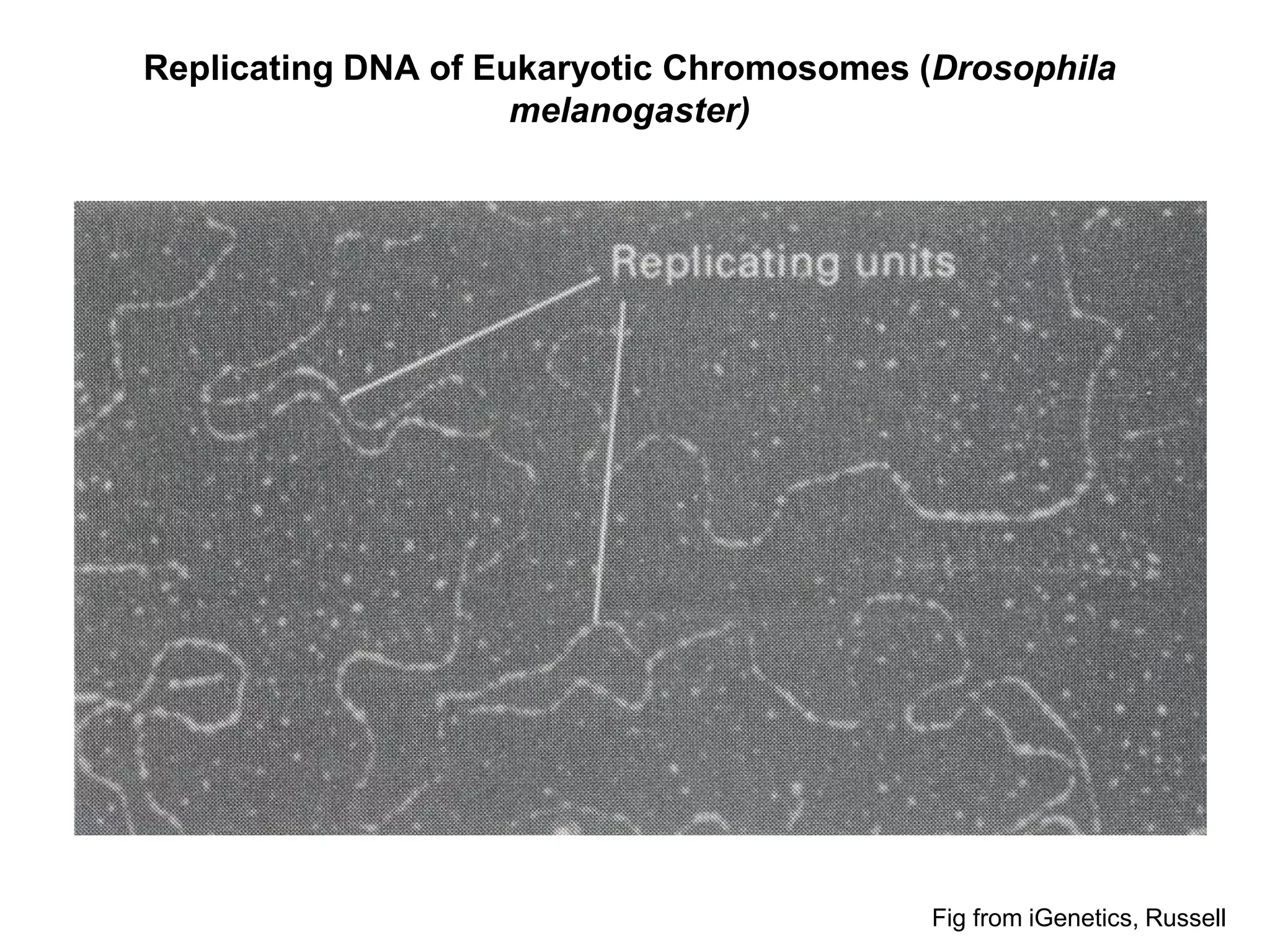
The document provides an overview of DNA replication in eukaryotes and prokaryotes. It discusses that eukaryotes have multiple origins of replication on their linear chromosomes compared to a single origin in prokaryotes. Eukaryotic DNA replication occurs more slowly than prokaryotes due to the complexity of unwinding DNA. The document also notes that linear chromosomes have difficulty replicating chromosome ends fully due to the inability of DNA polymerase to add nucleotides without a primer, posing problems for telomere maintenance with each cell division.











![Replication rate
• Eukaryotic DNA replication
– Typical human chromosome length: 100 million bp
– Time to replicate a chromosome: minutes to hours
– Hundreds of origins per chromosome
– Replicon = ~20,000 to 300,000 bp long
– 500-5000 bp / minute at each replication fork
(slower than bacterial replication; that much harder
to “unwind” the DNA for replication).
• Bacterial (prokaryotic) replication:
– Single circular chromosome (~4.6 million base pairs
[bp])
– Single origin of replication single replicon
(“Replication Bubble”)](https://image.slidesharecdn.com/dna-replication-221127223618-5cec010a/75/dna-replication-ppt-23-2048.jpg)