The document describes several key concepts in molecular biology:
1) The central dogma of gene expression describes how DNA is transcribed into RNA and then translated into protein. In bacteria this occurs in the cytoplasm, while in eukaryotes transcription occurs in the nucleus and translation in the cytosol.
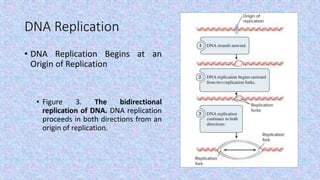
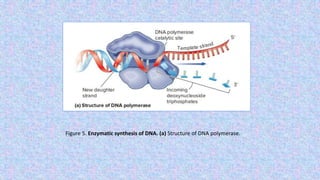
2) DNA replication is necessary for cell division and involves unwinding of the DNA double helix and synthesis of new strands according to the AT/GC base pairing rule, with the involvement of enzymes like DNA polymerase and helicase.
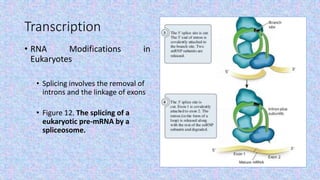
3) Transcription involves RNA polymerase using a DNA template to make RNA, and in eukaryotes the pre-mRNA undergoes splicing and capping/polyadenylation