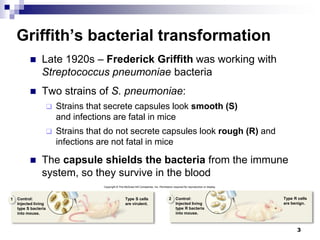
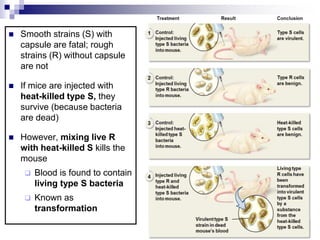
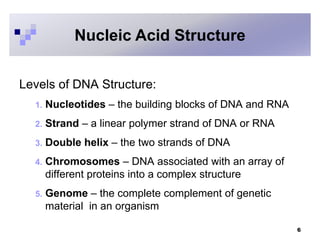
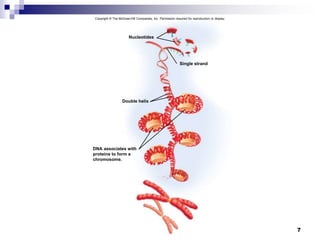
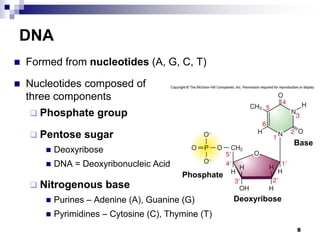
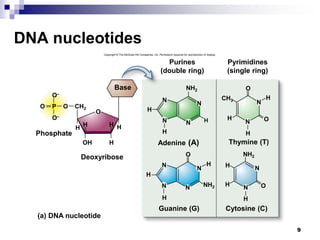
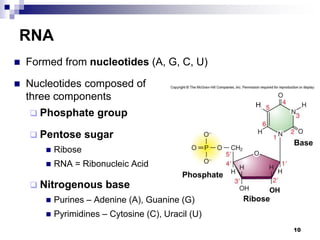
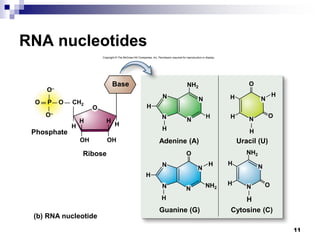
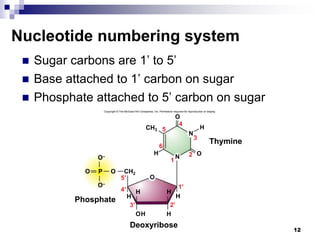
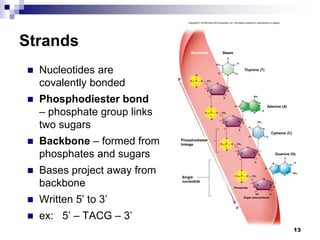
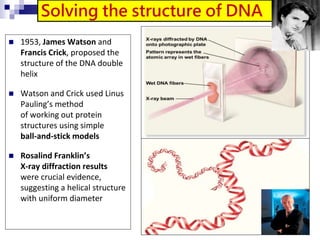
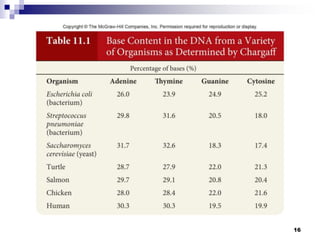
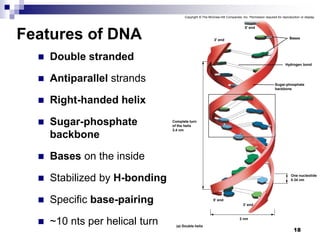
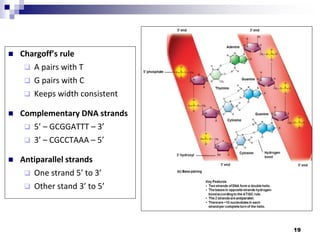
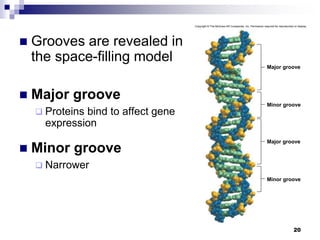
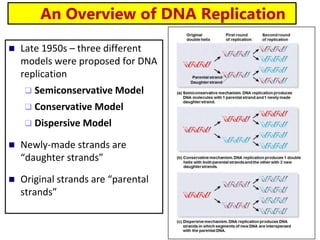
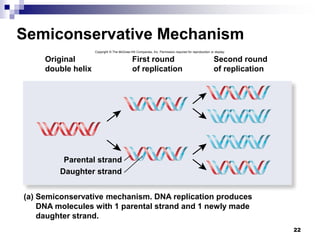
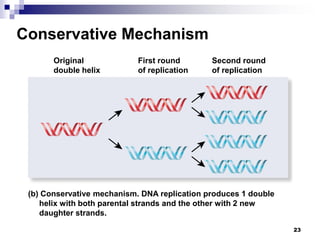
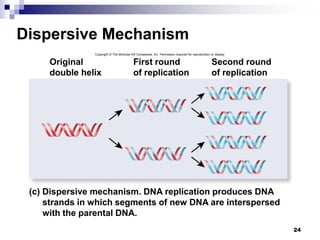
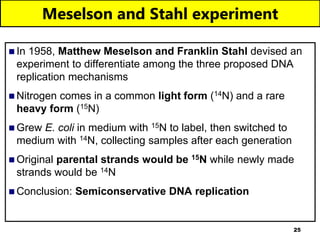
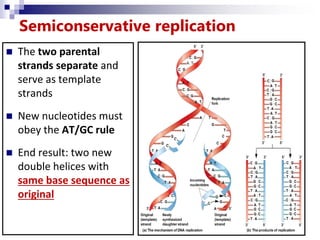
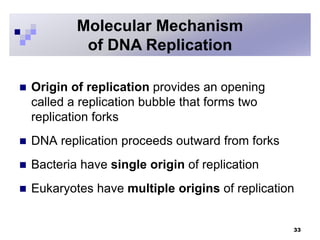
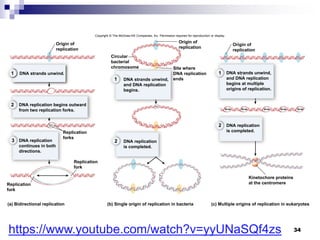
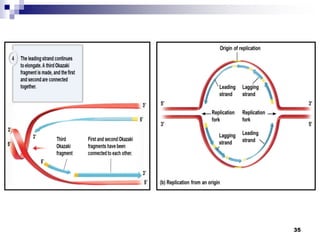
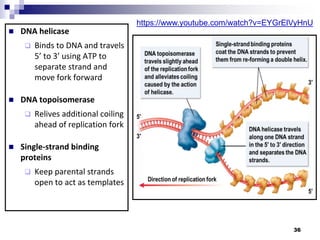
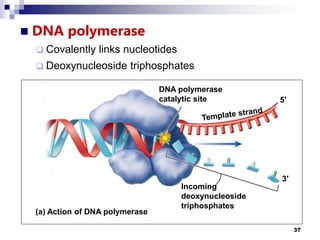
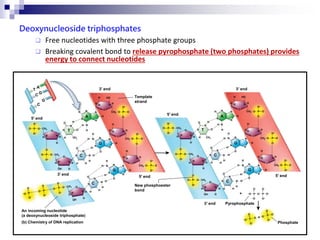
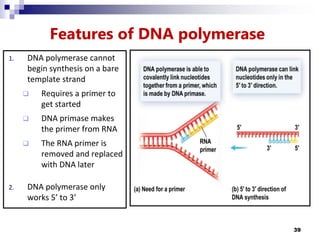
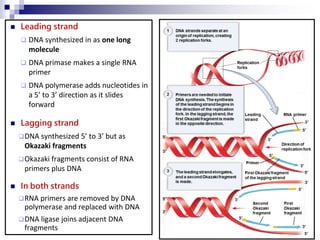
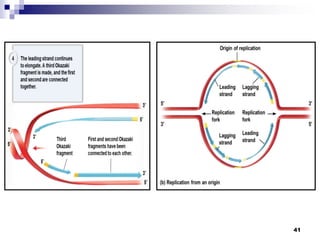
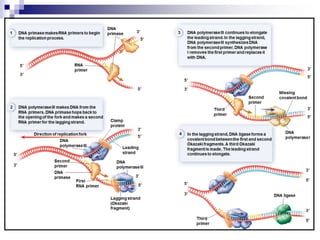
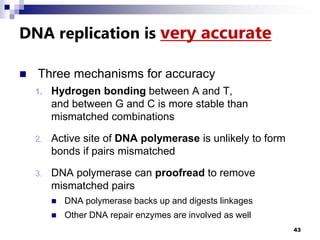
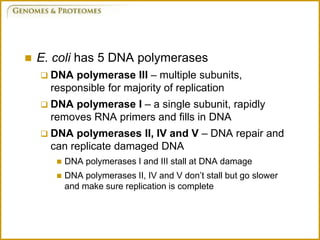
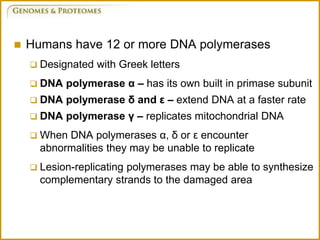
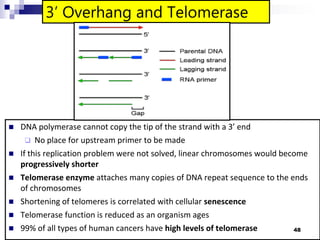
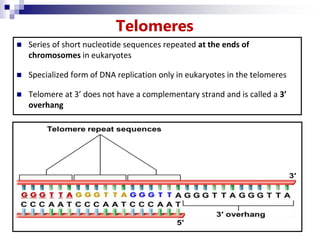
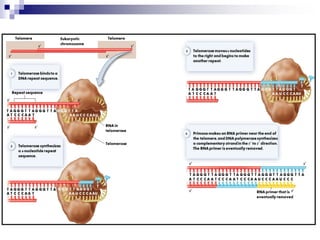
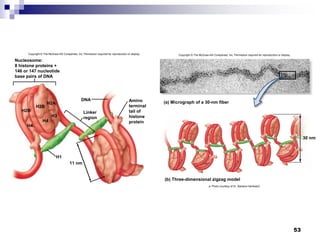
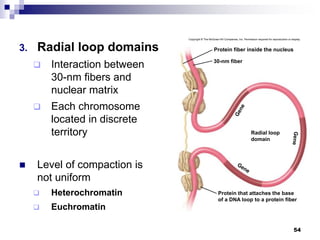
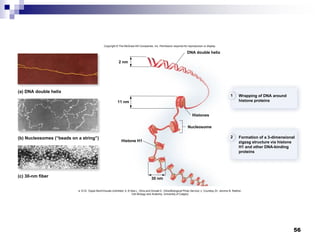
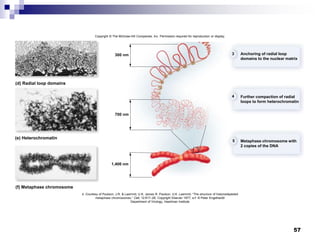
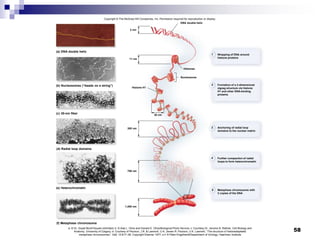
The document discusses DNA structure and replication. It begins by summarizing Griffith's experiments which showed genetic material can be transferred between bacteria. Next, it describes the discovery of DNA's double helix structure by Watson and Crick in 1953, including its key features like base pairing and antiparallel strands. The document then reviews three proposed models of DNA replication before summarizing Meselson and Stahl's experiment which supported the semiconservative model where each new DNA molecule contains one original and one new strand. Finally, it provides an overview of the molecular mechanism of DNA replication from the origin of replication to DNA polymerase adding nucleotides.