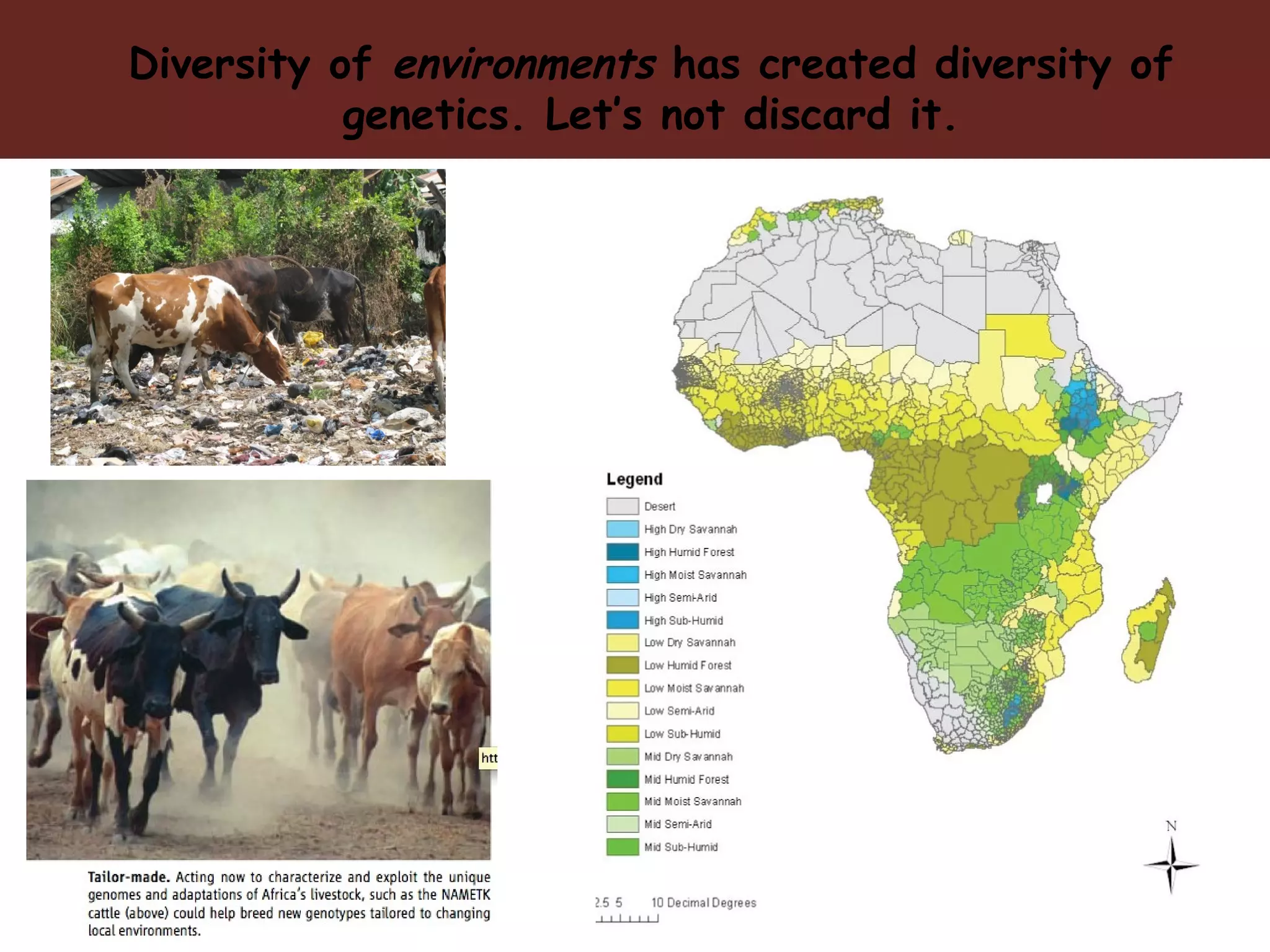
The document discusses the vision for livestock genetics in Africa, emphasizing the need for genetic gain while considering the diverse challenges faced by small-scale producers. Key issues include the impact of diseases like African trypanosomiasis on livestock and the limitations of current control methods such as vector control and vaccine development. It advocates for leveraging advancements in genome editing and data systems to identify and utilize natural genetic variations that improve livestock adaptation and productivity.