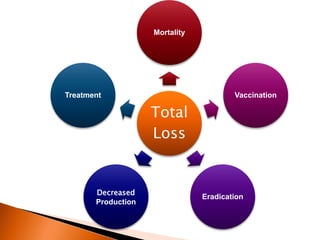
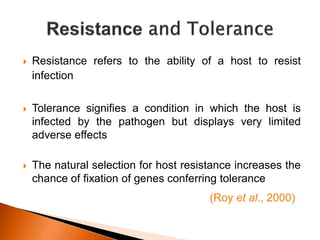
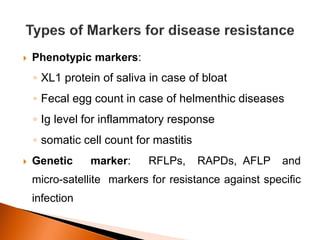
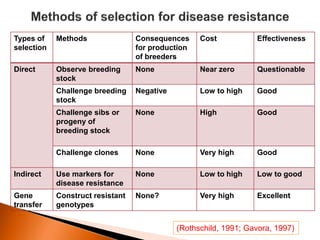
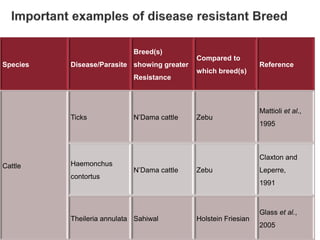
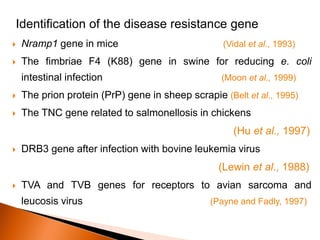
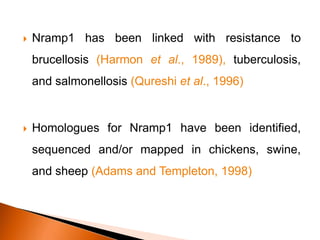
Dr. Jayesh Vyas discusses the impact of infectious diseases on the livestock industry, highlighting the economic losses and challenges in developing disease resistance through genetic selection. The document explores various methods, including direct and indirect selection for disease resistance, the identification of genetic markers linked to resistance traits, and the importance of integrating genetic approaches with management strategies. It emphasizes that while genetic selection can improve livestock health, it is not a standalone solution and must be accompanied by comprehensive management practices.