Genomic approaches to trypanosome resistance
•
1 like•48 views
Elucidating changes in gene expression in Tryp susceptible and resistant cattle during progression of tryp infection using Affymetrix gene expression Micro arrays
Report
Share
Report
Share
Download to read offline
Recommended
20100311 M Sc Lecture Final

supragenome presentation to Infection and Immunity MSc Class 11th March, 2010, Royal Free Hospital, London, UK
Functional profile of the pre- to post-mortem transition in blood

GTEx Project community meeting,
CRG, Barcelona, April 20th, 2017
A look at the human mutational load from the systems biology perspective

Molecular Evolution and Medicine Symposium
Temple University, Philadelphia, USA
16-17 September, 2017
Recommended
20100311 M Sc Lecture Final

supragenome presentation to Infection and Immunity MSc Class 11th March, 2010, Royal Free Hospital, London, UK
Functional profile of the pre- to post-mortem transition in blood

GTEx Project community meeting,
CRG, Barcelona, April 20th, 2017
A look at the human mutational load from the systems biology perspective

Molecular Evolution and Medicine Symposium
Temple University, Philadelphia, USA
16-17 September, 2017
Genome responses of trypanosome infected cattle

Genomic gene expression changes resulting from Trypanosomiasis: a horizontal study Examining expression changes elucidated by micro arrays in seminal tissues associated with the pathophysiology of Trypanosomiasis during disease progression
Bio380 Cancer Phylogenomics

Bio380 lecture on cancer as an evolutionary process, showing descent with modification, branching evolution and natural selection; focus on genome evolution
Synonymous mutations as drivers in human cancer genomes.

Synonymous mutations change the sequence of a gene without directly altering the sequence of the encoded protein. Here, we present evidence that these "silent" mutations frequently contribute to human cancer. Selection on synonymous mutations in oncogenes is cancer-type specific, and although the functional consequences of cancer-associated synonymous mutations may be diverse, they recurrently alter exonic motifs that regulate splicing and are associated with changes in oncogene splicing in tumors. The p53 tumor suppressor (TP53) also has recurrent synonymous mutations, but, in contrast to those in oncogenes, these are adjacent to splice sites and inactivate them. We estimate that between one in two and one in five silent mutations in oncogenes have been selected, equating to ~6%- 8% of all selected single-nucleotide changes in these genes. In addition, our analyses suggest that dosage-sensitive oncogenes have selected mutations in their 3' UTRs.
Multigenic (mechanistic) biomarkers

SEQC2, Advancing Precision Medicine and Cancer Genomics,
Bethesda, MD, September 13‐14, 2016
The interferons and their receptors – distribution and

The interferons and their receptors – distribution andGRUPO DE INVESTIGACION EN INMUNOLOGIA AREQUIPA
20091201 Transfer Seminar Final

MPhil->PhD transfer seminar to Infection and Immunity Department, 1st December, 2009, Windeyer Building, UCL, London, UK
Monocyte recruitment into atherosclerotic plaques

Monocyte recruitment into atherosclerotic plaquesSociety for Heart Attack Prevention and Eradication
SHAPE Society079 monocyte recruitment into atherosclerotic plaques

079 monocyte recruitment into atherosclerotic plaquesSociety for Heart Attack Prevention and Eradication
SHAPE SocietyBiochemistry Poster

Alpha-1 Antitrypsin (α-1 AT) deficiency is a common genetic disorder that affects 1 in 2,000 individuals in the USA. Additionally, over 20 million people have been identified as carriers for this genetic disorder. In severe cases, α-1 AT deficiency can cause substantial lung and liver damage, which if left untreated could result in death and there are no current available treatments. Alpha-1 protein is produced in the liver, travels in the bloodstream and utilized in the lungs to protect healthy lung tissue from harmful destruction by elastase. A common single amino acid substitution, located at E342K (ATZ) was identified in α-1 AT deficient humans. When this specific mutation occurs two phenotypes can result: 1) ATZ can polymerize in the liver causing cellular toxicity 2) inhibits alpha-1 antitrypsin from inhibiting elastase which can result in lung disease. Currently; little is known about the cellular mechanisms that clear the accumulated proteins in the liver. Therefore, an investigative study utilizing C. elegans model of ATZ was performed in order to help determine the cellular mechanisms that dispose of accumulated proteins. Specifically RNA interference was utilized to knockdown expression of specific genes. This investigation examined genes involved in the heat-shock pathway (HSP), unfolded protein response (UPR), and insulin signaling pathway (IS). Phenotypic analysis including: embryonic lethality, protein aggregation expression, and longevity, was completed after knockdown of genes to determine effect on ATZ accumulation. Currently with our preliminary data suggests that the heat-shack pathway may play a role in ATZ accumulation. Determining the mechanism of protein accumulation in the investigation of C. elegans may lead to possible drug targets and therefore the development of a treatment which may alleviate those diagnosed with this disorder.
Malaria disease 

Introduction of malaria
parasites responsible for malaria disease
primers for PCR
various stages of plasmodium falciparum infection
Systemic analysis of data combined from genetic qtl's and gene expression dat...

Systemic analysis of data combined from genetic qtl's and gene expression dat...Laurence Dawkins-Hall
Elucidating changes in gene expression by Micro array genomic sweeps of genetic QTLs linked to Tryp resistance in WT cattle to identify putative candidates underpinning pathophysiology Src jbbr-20-120 Dr. ihsan edan abdulkareem alsaimary PROFESSOR IN MEDICAL M...

Dr. ihsan edan abdulkareem alsaimary
PROFESSOR IN MEDICAL MICROBIOLOGY AND MOLECULAR IMMUNOLOGY
ihsanalsaimary@gmail.com
mobile : 009647801410838
university of basrah - college of medicine - basrah -IRAQ
Assessment of immunomolecular_expression_and_prognostic_role_of_tlr7_among_pa...

Dr. ihsan edan abdulkareem alsaimary
PROFESSOR IN MEDICAL MICROBIOLOGY AND MOLECULAR IMMUNOLOGY
ihsanalsaimary@gmail.com
mobile : 009647801410838
university of basrah - college of medicine - basrah -IRAQ
More Related Content
What's hot
Genome responses of trypanosome infected cattle

Genomic gene expression changes resulting from Trypanosomiasis: a horizontal study Examining expression changes elucidated by micro arrays in seminal tissues associated with the pathophysiology of Trypanosomiasis during disease progression
Bio380 Cancer Phylogenomics

Bio380 lecture on cancer as an evolutionary process, showing descent with modification, branching evolution and natural selection; focus on genome evolution
Synonymous mutations as drivers in human cancer genomes.

Synonymous mutations change the sequence of a gene without directly altering the sequence of the encoded protein. Here, we present evidence that these "silent" mutations frequently contribute to human cancer. Selection on synonymous mutations in oncogenes is cancer-type specific, and although the functional consequences of cancer-associated synonymous mutations may be diverse, they recurrently alter exonic motifs that regulate splicing and are associated with changes in oncogene splicing in tumors. The p53 tumor suppressor (TP53) also has recurrent synonymous mutations, but, in contrast to those in oncogenes, these are adjacent to splice sites and inactivate them. We estimate that between one in two and one in five silent mutations in oncogenes have been selected, equating to ~6%- 8% of all selected single-nucleotide changes in these genes. In addition, our analyses suggest that dosage-sensitive oncogenes have selected mutations in their 3' UTRs.
Multigenic (mechanistic) biomarkers

SEQC2, Advancing Precision Medicine and Cancer Genomics,
Bethesda, MD, September 13‐14, 2016
The interferons and their receptors – distribution and

The interferons and their receptors – distribution andGRUPO DE INVESTIGACION EN INMUNOLOGIA AREQUIPA
20091201 Transfer Seminar Final

MPhil->PhD transfer seminar to Infection and Immunity Department, 1st December, 2009, Windeyer Building, UCL, London, UK
Monocyte recruitment into atherosclerotic plaques

Monocyte recruitment into atherosclerotic plaquesSociety for Heart Attack Prevention and Eradication
SHAPE Society079 monocyte recruitment into atherosclerotic plaques

079 monocyte recruitment into atherosclerotic plaquesSociety for Heart Attack Prevention and Eradication
SHAPE SocietyBiochemistry Poster

Alpha-1 Antitrypsin (α-1 AT) deficiency is a common genetic disorder that affects 1 in 2,000 individuals in the USA. Additionally, over 20 million people have been identified as carriers for this genetic disorder. In severe cases, α-1 AT deficiency can cause substantial lung and liver damage, which if left untreated could result in death and there are no current available treatments. Alpha-1 protein is produced in the liver, travels in the bloodstream and utilized in the lungs to protect healthy lung tissue from harmful destruction by elastase. A common single amino acid substitution, located at E342K (ATZ) was identified in α-1 AT deficient humans. When this specific mutation occurs two phenotypes can result: 1) ATZ can polymerize in the liver causing cellular toxicity 2) inhibits alpha-1 antitrypsin from inhibiting elastase which can result in lung disease. Currently; little is known about the cellular mechanisms that clear the accumulated proteins in the liver. Therefore, an investigative study utilizing C. elegans model of ATZ was performed in order to help determine the cellular mechanisms that dispose of accumulated proteins. Specifically RNA interference was utilized to knockdown expression of specific genes. This investigation examined genes involved in the heat-shock pathway (HSP), unfolded protein response (UPR), and insulin signaling pathway (IS). Phenotypic analysis including: embryonic lethality, protein aggregation expression, and longevity, was completed after knockdown of genes to determine effect on ATZ accumulation. Currently with our preliminary data suggests that the heat-shack pathway may play a role in ATZ accumulation. Determining the mechanism of protein accumulation in the investigation of C. elegans may lead to possible drug targets and therefore the development of a treatment which may alleviate those diagnosed with this disorder.
Malaria disease 

Introduction of malaria
parasites responsible for malaria disease
primers for PCR
various stages of plasmodium falciparum infection
What's hot (19)
Regulatory b cells and tolerance in transplantation from animal models to hum...

Regulatory b cells and tolerance in transplantation from animal models to hum...
Synonymous mutations as drivers in human cancer genomes.

Synonymous mutations as drivers in human cancer genomes.
The interferons and their receptors – distribution and

The interferons and their receptors – distribution and
079 monocyte recruitment into atherosclerotic plaques

079 monocyte recruitment into atherosclerotic plaques
Similar to Genomic approaches to trypanosome resistance
Systemic analysis of data combined from genetic qtl's and gene expression dat...

Systemic analysis of data combined from genetic qtl's and gene expression dat...Laurence Dawkins-Hall
Elucidating changes in gene expression by Micro array genomic sweeps of genetic QTLs linked to Tryp resistance in WT cattle to identify putative candidates underpinning pathophysiology Src jbbr-20-120 Dr. ihsan edan abdulkareem alsaimary PROFESSOR IN MEDICAL M...

Dr. ihsan edan abdulkareem alsaimary
PROFESSOR IN MEDICAL MICROBIOLOGY AND MOLECULAR IMMUNOLOGY
ihsanalsaimary@gmail.com
mobile : 009647801410838
university of basrah - college of medicine - basrah -IRAQ
Assessment of immunomolecular_expression_and_prognostic_role_of_tlr7_among_pa...

Dr. ihsan edan abdulkareem alsaimary
PROFESSOR IN MEDICAL MICROBIOLOGY AND MOLECULAR IMMUNOLOGY
ihsanalsaimary@gmail.com
mobile : 009647801410838
university of basrah - college of medicine - basrah -IRAQ
A systematic, data driven approach to the combined analysis of microarray and...

A systematic, data driven approach to the combined analysis of microarray and...Laurence Dawkins-Hall
The use of gene expression data from Micro arrays coupled with WT QTL's linked to Tryp resistance phenotypes in Cattle to elucidate pertinent genetic changes underpinning phenotype in putative candidate genes Insights into the tumor microenvironment and therapeutic T cell manufacture r...

Insights into the tumor microenvironment and therapeutic T cell manufacture r...Thermo Fisher Scientific
TCRβ immune repertoire analysis by next-generation sequencing is emerging as a valuable tool for research studies of the tumor microenvironment and potential immune responses to cancer immunotherapy1-4. Here we describe a multiplex PCR-based TCRβ sequencing assay (Ion AmpliSeqTM Immune Repertoire Assay Plus – TCRβ) that leverages Ion AmpliSeq library construction chemistry and the long read capability of the Ion S5 530TM chip to provide coverage of all three CDR domains of the human TCRβ chain. We demonstrate use of the assay to evaluate tumor-infiltrating T cell repertoire features and monitor manufacture of therapeutic T cells.Proteomic Analysis of the Serum and Excretory-Secretary proteins of Trichinel...

The nematodes of the genus Trichinella are known to cause the pressing foodborne parasitic disease Trichinellosis and these parasites are known to complete all stages of development in one host with the enteral and parenteral phases observed during infection. Proteomics, in general, pertains to the systematic identification and quantification of the totality of proteins, which is the proteome of a biological system, at a specific point in time. The available proteomic studies have paved the way to identify and characterize Trichinella stage-specific proteins reacting with infected host-specific antibodies. Yet, very few contributions provide any information about changes in the global proteomic serum profile of Trichinella-infested individuals. Studies demonstrate that various Trichinella species and their phases of the invasion produce a characteristic proteomic pattern in the serum of experimentally infected pigs. Recent investigations have found that T. spiralis infection induced strong regulatory T cell responses through parasite excretory-secretory (ES) products, characterized by an increase of some regulatory T cells and growth factors. T. spiralis has also been reported to induce the angiogenic molecule vascular endothelial cell growth factor (VEGF) during nurse cell formation towards the induction of angiogenesis for nutrient supply and waste disposal. Herein, the various analogs considered in these studies include the serum, excretory-secretory proteins, surface proteins, immune-reactive proteins from muscle larvae (ML) and so on. Intestinal cultures, striated muscle tissues, pigs, mice, beavers, contributions from patients are some of the major models exploited for this purpose. The current analysis focuses on recapitulating the recent findings driven on this area to create a common ground for further studies and to ease any difficulty in continuing the proteomic analysis of T. spiralis using in vitro and in vivo models.
characterization of FQ Non-susceptible S. Pyogenes

The aim of this study was to investigate the frequency, mechanism, and epidemiological association of FQ non susceptibility in S.pyogenes during 2011 and 2016 from Shanghai, China.
identification and characterization of FQ-non-susceptable S. Pyogenes

The aim of this study was to investigate the frequency, mechanism, and epidemiological association of FQ non susceptibility in S.pyogenes during 2011 and 2016 from Shanghai, China.
The 'omics' revolution: How will it improve our understanding of infections a...

This slideset explains the ‘Omics’ technology and its role in the study of infections and vaccination. It is a revolution as it offers powerful tools to interrogate the animal / human immune response to vaccines and infections.
Shigella flexneri serotype 1c derived from serotype 1a by acquisition of gtrI...

Background
Shigella spp. are the primary causative agents of bacillary dysentery. Since its emergence in the late 1980s, the S. flexneri serotype 1c remains poorly understood, particularly with regard to its origin and genetic evolution. This article provides a molecular insight into this novel serotype and the gtrIC gene cluster that determines its unique immune recognition.
Results
A PCR of the gtrIC cluster showed that serotype 1c isolates from different geographical origins were genetically conserved. An analysis of sequences flanking the gtrIC cluster revealed remnants of a prophage genome, in particular integrase and tRNAPro genes. Meanwhile, Southern blot analyses on serotype 1c, 1a and 1b strains indicated that all the tested serotype 1c strains may have had a common origin that has since remained distinct from the closely related 1a and 1b serotypes. The identification of prophage genes upstream of the gtrIC cluster is consistent with the notion of bacteriophage-mediated integration of the gtrIC cluster into a pre-existing serotype.
Conclusions
This is the first study to show that serotype 1c isolates from different geographical origins share an identical pattern of genetic arrangement, suggesting that serotype 1c strains may have originated from a single parental strain. Analysis of the sequence around the gtrIC cluster revealed a new site for the integration of the serotype converting phages of S. flexneri. Understanding the origin of new pathogenic serotypes and the molecular basis of serotype conversion in S. flexneri would provide information for developing cross-reactive Shigella vaccines.
Keywords
Shigella flexneri, Bacillary dysentery, Serotype-conversion , Evolutionary origin, Glucosyltransferase, Serotype 1c
Ceraseetal2021complete.pdf

Female mammals achieve dosage compensation by inactivating one of their two X chromosomes
during development, a process entirely dependent on Xist, an X-linked long noncoding
RNA (lncRNA). At the onset of X chromosome inactivation (XCI), Xist is up-regulated
and spreads along the future inactive X chromosome. Contextually, it recruits repressive
histone and DNA modifiers that transcriptionally silence the X chromosome. Xist regulation is
tightly coupled to differentiation and its expression is under the control of both pluripotency
and epigenetic factors. Recent evidence has suggested that chromatin remodelers accumulate
at the X Inactivation Center (XIC) and here we demonstrate a new role for Chd8 in Xist
regulation in differentiating ES cells, linked to its control and prevention of spurious
transcription factor interactions occurring within Xist regulatory regions. Our findings have a
broader relevance, in the context of complex, developmentally-regulated gene expression.
Similar to Genomic approaches to trypanosome resistance (20)
Systemic analysis of data combined from genetic qtl's and gene expression dat...

Systemic analysis of data combined from genetic qtl's and gene expression dat...
Src jbbr-20-120 Dr. ihsan edan abdulkareem alsaimary PROFESSOR IN MEDICAL M...

Src jbbr-20-120 Dr. ihsan edan abdulkareem alsaimary PROFESSOR IN MEDICAL M...
Assessment of immunomolecular_expression_and_prognostic_role_of_tlr7_among_pa...

Assessment of immunomolecular_expression_and_prognostic_role_of_tlr7_among_pa...
A systematic, data driven approach to the combined analysis of microarray and...

A systematic, data driven approach to the combined analysis of microarray and...
Insights into the tumor microenvironment and therapeutic T cell manufacture r...

Insights into the tumor microenvironment and therapeutic T cell manufacture r...
Bioinformatics-driven discovery of EGFR mutant Lung Cancer

Bioinformatics-driven discovery of EGFR mutant Lung Cancer
Proteomic Analysis of the Serum and Excretory-Secretary proteins of Trichinel...

Proteomic Analysis of the Serum and Excretory-Secretary proteins of Trichinel...
characterization of FQ Non-susceptible S. Pyogenes

characterization of FQ Non-susceptible S. Pyogenes
identification and characterization of FQ-non-susceptable S. Pyogenes

identification and characterization of FQ-non-susceptable S. Pyogenes
The 'omics' revolution: How will it improve our understanding of infections a...

The 'omics' revolution: How will it improve our understanding of infections a...
Diversity of O Antigens within the Genus Cronobacter - Martina

Diversity of O Antigens within the Genus Cronobacter - Martina
Shigella flexneri serotype 1c derived from serotype 1a by acquisition of gtrI...

Shigella flexneri serotype 1c derived from serotype 1a by acquisition of gtrI...
More from Laurence Dawkins-Hall
DSRG report 2001

Describing the activities of the DNA Sequencing Research group (DSRG) of the ABRF for the year 2001:
Journal of Bio molecular Techniques 12:27–29 © 2001 ABRF
An evaluation of methods used to sequence pGEM template within core facilitie...

An evaluation of methods used to sequence pGEM template within core facilitie...Laurence Dawkins-Hall
A horizontal study by ABRF designed to investigate & collate cycle sequencing methods for the pGEM template and subsequent data output on automated sequencing platforms (cf. AB 3700 & AB 3100) 06 isafg p77 genome wide expression consequences of a disease resistance qtl ...

06 isafg p77 genome wide expression consequences of a disease resistance qtl ...Laurence Dawkins-Hall
Presentation describing the genetic pathophysiology of Trypanosomiasis resistance in WT cattleLab talk 181209 radioligand assay for validating in silico rel 1 antagonist h...

Lab talk 181209 radioligand assay for validating in silico rel 1 antagonist h...Laurence Dawkins-Hall
Titrating preliminary hits derived from anti REL 1 radio mimetic assay: Precursor to full regression analysis of lead compounds to establish IC50'sLab talk 201109 radioligand assay for validating in slico predicted rel1 anta...

Lab talk 201109 radioligand assay for validating in slico predicted rel1 anta...Laurence Dawkins-Hall
preliminary quantification of radio mimetic anti REL 1 hits by titration: Scoping likely IC50Lab talk 180909 radioligand assay to validate in silico predicted rel1 inhib...

Lab talk 180909 radioligand assay to validate in silico predicted rel1 inhib...Laurence Dawkins-Hall
implementation of a radio ligand assay to validate antagonists of the Tryp RNA editing enzyme REL 1 to validate in silico predictionsLab talk 060809 radioligand assay to validate in slico predicted rel inhibito...

Lab talk 060809 radioligand assay to validate in slico predicted rel inhibito...Laurence Dawkins-Hall
Radio ligand assay to validate putative antagonists of REL1 predicted by in silico modellingLabtalk 101210 optimising fret assay_den_optimising r_rel 1 production_heat s...

Labtalk 101210 optimising fret assay_den_optimising r_rel 1 production_heat s...Laurence Dawkins-Hall
optimising HT FRET drug screening assay with ref to denaturation regimen; potentiation of soluble REL 1 production by heat shock inclusion Labtalk 151010 optimising fret assay_ligation buffer_[atp]_fluorophores![Labtalk 151010 optimising fret assay_ligation buffer_[atp]_fluorophores](data:image/gif;base64,R0lGODlhAQABAIAAAAAAAP///yH5BAEAAAAALAAAAAABAAEAAAIBRAA7)
![Labtalk 151010 optimising fret assay_ligation buffer_[atp]_fluorophores](data:image/gif;base64,R0lGODlhAQABAIAAAAAAAP///yH5BAEAAAAALAAAAAABAAEAAAIBRAA7)
Optimising parameters associated with a high through put FRET assay for identifying Tryp REL 1 antagonists, viz.
1. Ligation conditions, cf. buffer composition e.g. pH; ligation time etc
2. Mixing and matching fluorophores to maximise S:N and hence assay sensitivity
Lab talk 300710 optimisation of fret assay parameters_calculating km of atp s...

Lab talk 300710 optimisation of fret assay parameters_calculating km of atp s...Laurence Dawkins-Hall
Optimisation of FRET parameters to improve FRET compound screening assay, viz
1. Denaturation regimen
2. Establishing Km of [ATP] substrateLab talk 070510 inducing r_rel1 for establishing fret assay_ic50 studies with...

Lab talk 070510 inducing r_rel1 for establishing fret assay_ic50 studies with...Laurence Dawkins-Hall
1. more work at optimising expression and purification of rREL 1 for incorporation into a high throughput FRET assay for compound library screens to identify REL 1 antagonists
2. Regression analysis of titrants of compounds identified as demonstrating REL 1 inhibition by radiomimetic assay; quantification of efficacy by means of IC50 Lab talk 020410 inducing rel 1 to set up ht fret assay_progress

Outlining procedural pipeline for expressing and purifying active rREL 1 for incorporation into high throughput fluorescent (FRET) screening assay for identifying antagonistic hits of REL 1
Lab talk 190210 efficacy studies on radioligand hits_beginnings of fret assay...

Lab talk 190210 efficacy studies on radioligand hits_beginnings of fret assay...Laurence Dawkins-Hall
1. Titration of REL 1 antagonist hits identified by radio mimetic assay to quantify efficacy (IC50)
2. Exposition of HT FRET assay principles for large scale compound library screens to empirically identify REL 1 antagonist hitsLab talk 020710 comparing bac r_rel 1 with e coli rrel 1 for use in fret assay

Comparing activity of baculovirus and E Coli expressed rREL 1 fractions in context of HT FRET assay for screening compound libraries to identify REL1 antagonist hits
Genomic analysis of a hypertensive qtl on rat chr 1

Transcriptomic analysis of rat QTL linked to hypertension by exomic sequencing and micro array gene expression analysis
RNA editing as a drug target in tryp assay dev woodshole 2011

Aspects of HT assay development for identifying putative inhibitors of the Tryp RNA editing enzyme REL 1 by compound library screens
RNA editing as a drug target in tryp assay dev woods_hole2_0411 acs v2

Aspects of HT assay development for identifying putative inhibitors of the Tryp RNA editing enzyme REL 1 by compound library screens
RNA editing as a drug target in tryp. development of a high throughput sceeni...

RNA editing as a drug target in tryp. development of a high throughput sceeni...Laurence Dawkins-Hall
Aspects of development of an HT assay for identifying inhibitors of the Tryp target protein REL 1: Potential microbial therapeutics by 'hit to lead'Rna editing as a drug target identification of inhibitors of rel 1 bsp 2010

Low through put radiomimetic assay to identify putative inhibitors of the Tryp enzyme REL 1 'in vitro' predicted by 'in silico' modelling
Rna editing as a drug target identification of inhibitors of rel 1 bsp 210

Low through put radiomimetic assay to identify putative inhibitors of the Tryp enzyme REL 1 'in vitro' predicted by 'in silico' modelling
More from Laurence Dawkins-Hall (20)
An evaluation of methods used to sequence pGEM template within core facilitie...

An evaluation of methods used to sequence pGEM template within core facilitie...
06 isafg p77 genome wide expression consequences of a disease resistance qtl ...

06 isafg p77 genome wide expression consequences of a disease resistance qtl ...
Lab talk 181209 radioligand assay for validating in silico rel 1 antagonist h...

Lab talk 181209 radioligand assay for validating in silico rel 1 antagonist h...
Lab talk 201109 radioligand assay for validating in slico predicted rel1 anta...

Lab talk 201109 radioligand assay for validating in slico predicted rel1 anta...
Lab talk 180909 radioligand assay to validate in silico predicted rel1 inhib...

Lab talk 180909 radioligand assay to validate in silico predicted rel1 inhib...
Lab talk 060809 radioligand assay to validate in slico predicted rel inhibito...

Lab talk 060809 radioligand assay to validate in slico predicted rel inhibito...
Labtalk 101210 optimising fret assay_den_optimising r_rel 1 production_heat s...

Labtalk 101210 optimising fret assay_den_optimising r_rel 1 production_heat s...
Labtalk 151010 optimising fret assay_ligation buffer_[atp]_fluorophores![Labtalk 151010 optimising fret assay_ligation buffer_[atp]_fluorophores](data:image/gif;base64,R0lGODlhAQABAIAAAAAAAP///yH5BAEAAAAALAAAAAABAAEAAAIBRAA7)
![Labtalk 151010 optimising fret assay_ligation buffer_[atp]_fluorophores](data:image/gif;base64,R0lGODlhAQABAIAAAAAAAP///yH5BAEAAAAALAAAAAABAAEAAAIBRAA7)
Labtalk 151010 optimising fret assay_ligation buffer_[atp]_fluorophores
Lab talk 300710 optimisation of fret assay parameters_calculating km of atp s...

Lab talk 300710 optimisation of fret assay parameters_calculating km of atp s...
Lab talk 070510 inducing r_rel1 for establishing fret assay_ic50 studies with...

Lab talk 070510 inducing r_rel1 for establishing fret assay_ic50 studies with...
Lab talk 020410 inducing rel 1 to set up ht fret assay_progress

Lab talk 020410 inducing rel 1 to set up ht fret assay_progress
Lab talk 190210 efficacy studies on radioligand hits_beginnings of fret assay...

Lab talk 190210 efficacy studies on radioligand hits_beginnings of fret assay...
Lab talk 020710 comparing bac r_rel 1 with e coli rrel 1 for use in fret assay

Lab talk 020710 comparing bac r_rel 1 with e coli rrel 1 for use in fret assay
Genomic analysis of a hypertensive qtl on rat chr 1

Genomic analysis of a hypertensive qtl on rat chr 1
RNA editing as a drug target in tryp assay dev woodshole 2011

RNA editing as a drug target in tryp assay dev woodshole 2011
RNA editing as a drug target in tryp assay dev woods_hole2_0411 acs v2

RNA editing as a drug target in tryp assay dev woods_hole2_0411 acs v2
RNA editing as a drug target in tryp. development of a high throughput sceeni...

RNA editing as a drug target in tryp. development of a high throughput sceeni...
Rna editing as a drug target identification of inhibitors of rel 1 bsp 2010

Rna editing as a drug target identification of inhibitors of rel 1 bsp 2010
Rna editing as a drug target identification of inhibitors of rel 1 bsp 210

Rna editing as a drug target identification of inhibitors of rel 1 bsp 210
Recently uploaded
Unveiling the Energy Potential of Marshmallow Deposits.pdf

Unveiling the Energy Potential of Marshmallow Deposits: A Revolutionary
Breakthrough in Sustainable Energy Science
THE IMPORTANCE OF MARTIAN ATMOSPHERE SAMPLE RETURN.

The return of a sample of near-surface atmosphere from Mars would facilitate answers to several first-order science questions surrounding the formation and evolution of the planet. One of the important aspects of terrestrial planet formation in general is the role that primary atmospheres played in influencing the chemistry and structure of the planets and their antecedents. Studies of the martian atmosphere can be used to investigate the role of a primary atmosphere in its history. Atmosphere samples would also inform our understanding of the near-surface chemistry of the planet, and ultimately the prospects for life. High-precision isotopic analyses of constituent gases are needed to address these questions, requiring that the analyses are made on returned samples rather than in situ.
原版制作(carleton毕业证书)卡尔顿大学毕业证硕士文凭原版一模一样

原版纸张【微信:741003700 】【(carleton毕业证书)卡尔顿大学毕业证】【微信:741003700 】学位证,留信认证(真实可查,永久存档)offer、雅思、外壳等材料/诚信可靠,可直接看成品样本,帮您解决无法毕业带来的各种难题!外壳,原版制作,诚信可靠,可直接看成品样本。行业标杆!精益求精,诚心合作,真诚制作!多年品质 ,按需精细制作,24小时接单,全套进口原装设备。十五年致力于帮助留学生解决难题,包您满意。
本公司拥有海外各大学样板无数,能完美还原海外各大学 Bachelor Diploma degree, Master Degree Diploma
1:1完美还原海外各大学毕业材料上的工艺:水印,阴影底纹,钢印LOGO烫金烫银,LOGO烫金烫银复合重叠。文字图案浮雕、激光镭射、紫外荧光、温感、复印防伪等防伪工艺。材料咨询办理、认证咨询办理请加学历顾问Q/微741003700
留信网认证的作用:
1:该专业认证可证明留学生真实身份
2:同时对留学生所学专业登记给予评定
3:国家专业人才认证中心颁发入库证书
4:这个认证书并且可以归档倒地方
5:凡事获得留信网入网的信息将会逐步更新到个人身份内,将在公安局网内查询个人身份证信息后,同步读取人才网入库信息
6:个人职称评审加20分
7:个人信誉贷款加10分
8:在国家人才网主办的国家网络招聘大会中纳入资料,供国家高端企业选择人才
Salas, V. (2024) "John of St. Thomas (Poinsot) on the Science of Sacred Theol...

I Introduction
II Subalternation and Theology
III Theology and Dogmatic Declarations
IV The Mixed Principles of Theology
V Virtual Revelation: The Unity of Theology
VI Theology as a Natural Science
VII Theology’s Certitude
VIII Conclusion
Notes
Bibliography
All the contents are fully attributable to the author, Doctor Victor Salas. Should you wish to get this text republished, get in touch with the author or the editorial committee of the Studia Poinsotiana. Insofar as possible, we will be happy to broker your contact.
Richard's aventures in two entangled wonderlands

Since the loophole-free Bell experiments of 2020 and the Nobel prizes in physics of 2022, critics of Bell's work have retreated to the fortress of super-determinism. Now, super-determinism is a derogatory word - it just means "determinism". Palmer, Hance and Hossenfelder argue that quantum mechanics and determinism are not incompatible, using a sophisticated mathematical construction based on a subtle thinning of allowed states and measurements in quantum mechanics, such that what is left appears to make Bell's argument fail, without altering the empirical predictions of quantum mechanics. I think however that it is a smoke screen, and the slogan "lost in math" comes to my mind. I will discuss some other recent disproofs of Bell's theorem using the language of causality based on causal graphs. Causal thinking is also central to law and justice. I will mention surprising connections to my work on serial killer nurse cases, in particular the Dutch case of Lucia de Berk and the current UK case of Lucy Letby.
Deep Software Variability and Frictionless Reproducibility

Deep Software Variability and Frictionless ReproducibilityUniversity of Rennes, INSA Rennes, Inria/IRISA, CNRS
The ability to recreate computational results with minimal effort and actionable metrics provides a solid foundation for scientific research and software development. When people can replicate an analysis at the touch of a button using open-source software, open data, and methods to assess and compare proposals, it significantly eases verification of results, engagement with a diverse range of contributors, and progress. However, we have yet to fully achieve this; there are still many sociotechnical frictions.
Inspired by David Donoho's vision, this talk aims to revisit the three crucial pillars of frictionless reproducibility (data sharing, code sharing, and competitive challenges) with the perspective of deep software variability.
Our observation is that multiple layers — hardware, operating systems, third-party libraries, software versions, input data, compile-time options, and parameters — are subject to variability that exacerbates frictions but is also essential for achieving robust, generalizable results and fostering innovation. I will first review the literature, providing evidence of how the complex variability interactions across these layers affect qualitative and quantitative software properties, thereby complicating the reproduction and replication of scientific studies in various fields.
I will then present some software engineering and AI techniques that can support the strategic exploration of variability spaces. These include the use of abstractions and models (e.g., feature models), sampling strategies (e.g., uniform, random), cost-effective measurements (e.g., incremental build of software configurations), and dimensionality reduction methods (e.g., transfer learning, feature selection, software debloating).
I will finally argue that deep variability is both the problem and solution of frictionless reproducibility, calling the software science community to develop new methods and tools to manage variability and foster reproducibility in software systems.
Exposé invité Journées Nationales du GDR GPL 2024
Orion Air Quality Monitoring Systems - CWS

Professional air quality monitoring systems provide immediate, on-site data for analysis, compliance, and decision-making.
Monitor common gases, weather parameters, particulates.
Nutraceutical market, scope and growth: Herbal drug technology

As consumer awareness of health and wellness rises, the nutraceutical market—which includes goods like functional meals, drinks, and dietary supplements that provide health advantages beyond basic nutrition—is growing significantly. As healthcare expenses rise, the population ages, and people want natural and preventative health solutions more and more, this industry is increasing quickly. Further driving market expansion are product formulation innovations and the use of cutting-edge technology for customized nutrition. With its worldwide reach, the nutraceutical industry is expected to keep growing and provide significant chances for research and investment in a number of categories, including vitamins, minerals, probiotics, and herbal supplements.
Comparing Evolved Extractive Text Summary Scores of Bidirectional Encoder Rep...

Comparing Evolved Extractive Text Summary Scores of Bidirectional Encoder Rep...University of Maribor
Slides from:
11th International Conference on Electrical, Electronics and Computer Engineering (IcETRAN), Niš, 3-6 June 2024
Track: Artificial Intelligence
https://www.etran.rs/2024/en/home-english/Recently uploaded (20)
Lateral Ventricles.pdf very easy good diagrams comprehensive

Lateral Ventricles.pdf very easy good diagrams comprehensive
BLOOD AND BLOOD COMPONENT- introduction to blood physiology

BLOOD AND BLOOD COMPONENT- introduction to blood physiology
Unveiling the Energy Potential of Marshmallow Deposits.pdf

Unveiling the Energy Potential of Marshmallow Deposits.pdf
THE IMPORTANCE OF MARTIAN ATMOSPHERE SAMPLE RETURN.

THE IMPORTANCE OF MARTIAN ATMOSPHERE SAMPLE RETURN.
Salas, V. (2024) "John of St. Thomas (Poinsot) on the Science of Sacred Theol...

Salas, V. (2024) "John of St. Thomas (Poinsot) on the Science of Sacred Theol...
Deep Software Variability and Frictionless Reproducibility

Deep Software Variability and Frictionless Reproducibility
In silico drugs analogue design: novobiocin analogues.pptx

In silico drugs analogue design: novobiocin analogues.pptx
Nutraceutical market, scope and growth: Herbal drug technology

Nutraceutical market, scope and growth: Herbal drug technology
Comparing Evolved Extractive Text Summary Scores of Bidirectional Encoder Rep...

Comparing Evolved Extractive Text Summary Scores of Bidirectional Encoder Rep...
PRESENTATION ABOUT PRINCIPLE OF COSMATIC EVALUATION

PRESENTATION ABOUT PRINCIPLE OF COSMATIC EVALUATION
Genomic approaches to trypanosome resistance
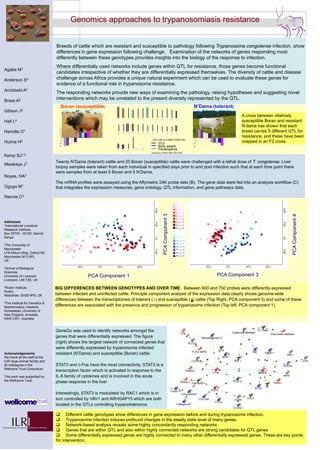
- 1. BIG DIFFERENCES BETWEEN GENOTYPES AND OVER TIME. Between 600 and 750 probes were differently expressed between infected and uninfected cattle. Principle component analysis of the expression data clearly shows genome-wide differences between the transcriptomes of tolerant (×) and susceptible ( ) cattle (Top Right, PCA component 3) and some of these differences are associated with the presence and progression of trypanosome infection (Top left, PCA component 1). This image cannot currently be displayed. This image cannot currently be displayed. Genomics approaches to trypanosomiasis resistance Breeds of cattle which are resistant and susceptible to pathology following Trypanosoma congolense infection, show differences in gene expression following challenge. Examination of the networks of genes responding most differently between these genotypes provides insights into the biology of the response to infection. Where differentially used networks include genes within QTL for resistance, those genes become functional candidates irrespective of whether they are differentially expressed themselves. The diversity of cattle and disease challenge across Africa provides a unique natural experiment which can be used to evaluate these genes for evidence of a functional role in trypanosome resistance. The responding networks provide new ways of examining the pathology, raising hypotheses and suggesting novel interventions which may be unrelated to the present diversity represented by the QTL. GeneGo was used to identify networks amongst the genes that were differentially expressed. The figure (right) shows the largest network of connected genes that were differently expressed by trypanosome infected resistant (N’Dama) and susceptible (Boran) cattle. STAT3 and c-Fos have the most connectivity. STAT3 is a transcription factor which is activated in response to the IL-6 family of cytokines and is involved in the acute phase response in the liver Interestingly, STAT3 is modulated by RAC1 which is in turn controlled by VAV1 and ARHGAP15 which are both located in the QTLs controlling trypanotolerance. Different cattle genotypes show differences in gene expression before and during trypanosome infection. Trypanosome infection induces profound changes in the steady state level of many genes. Network-based analysis reveals some highly concordantly responding networks Genes that are within QTL and also within highly connected networks are strong candidates for QTL genes Some differentially expressed genes are highly connected to many other differentially expressed genes. These are key points for intervention. Twenty N’Dama (tolerant) cattle and 20 Boran (susceptible) cattle were challenged with a lethal dose of T. congolense. Liver biopsy samples were taken from each individual in specified days prior to and post infection such that at each time point there were samples from at least 5 Boran and 5 N’Dama. The mRNA profiles were assayed using the Affymetrix 24K probe sets (B). The gene data were fed into an analysis workflow (C) that integrates the expression measures, gene ontology, QTL information, and gene pathways data. Agaba M1 Anderson S4 Archibald A4 Brass A2 Gibson J5 Hall L4 Hanotte O1 Hulme H2 Kemp SJ1,3 Mwakaya J1 Noyes, HA3 Ogugo M1 Rennie C3 Addresses 1International Livestock Research Institute, Box 30709 - 00100, Nairobi Kenya 2The University of Manchester LF8 Kilburn Bldg, Oxford Rd Manchester M13 9PL UK 3School of Biological Sciences, University of Liverpool, Liverpool, L69 7ZB, UK 4Roslin Institute, Roslin, Midlothian, EH25 9PS, UK 5The Institute for Genetics & Bioinformatics, Hawkins Homestead, University of New England, Armidale, NSW 2351, Australia Acknowledgements: We thank all the staff at the ILRI large animal facility and all colleagues in the Welcome Trust Consortium This work was supported by the Wellcome Trust. QTL with p<0.0043 FDR(10%) PCV Body weight Parasitaemia Hanotte et al PNAS 2003 7443-7448 -15 -10 -5 0 5 10 15 N’Dama (tolerant) -15 -10 -5 0 5 10 15 Boran (susceptible) A cross between relatively susceptible Boran and resistant N’dama has shown that each breed carries 5 different QTL for resistance, and these have been mapped in an F2 cross.
