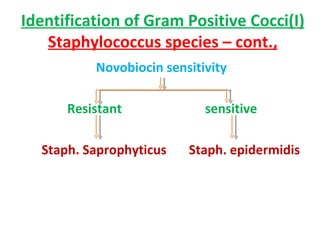
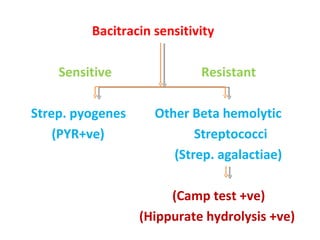
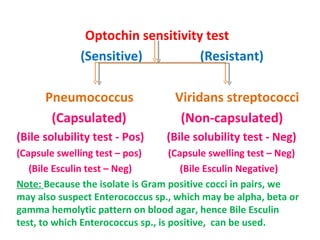
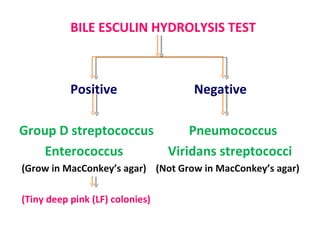
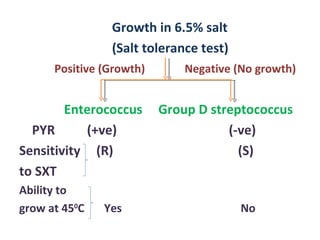
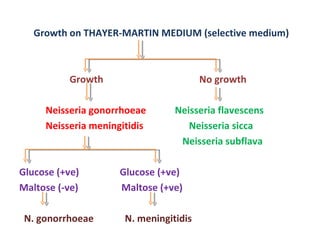
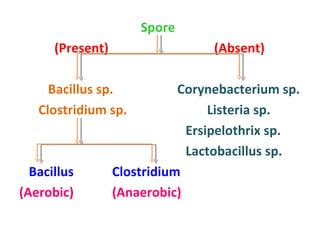
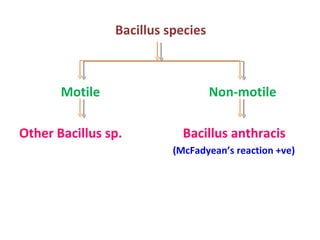
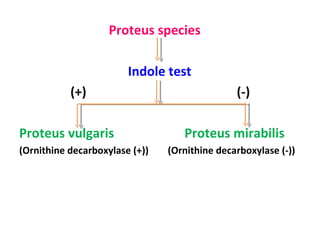
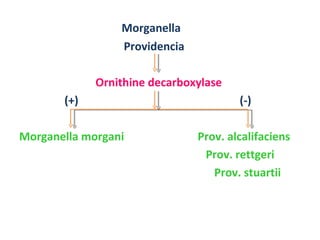
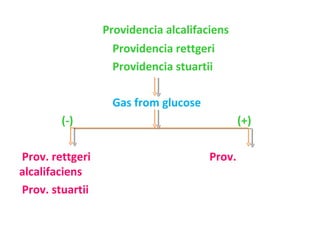
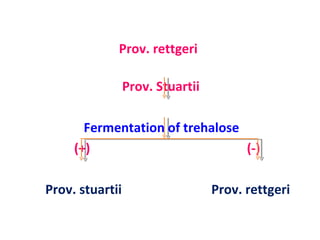
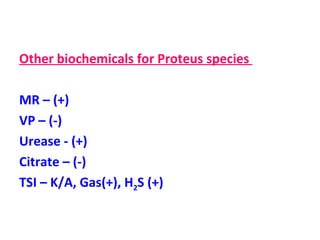
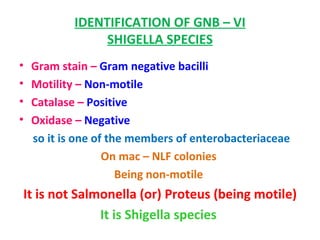
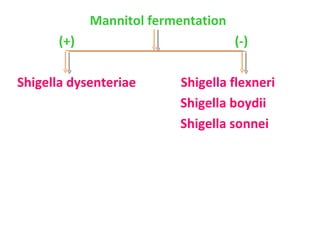
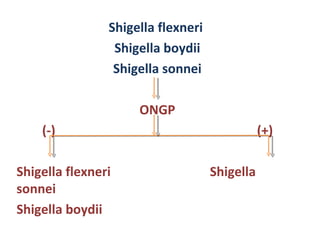
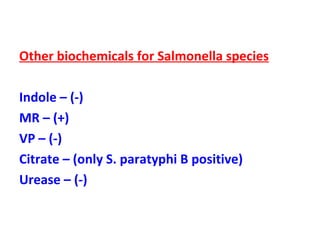
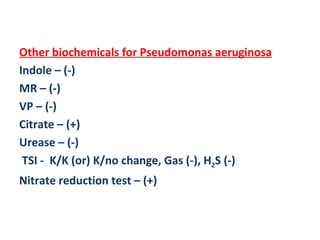
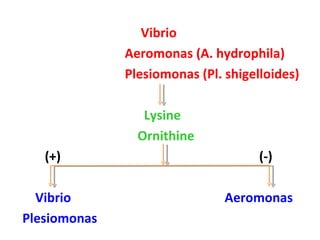
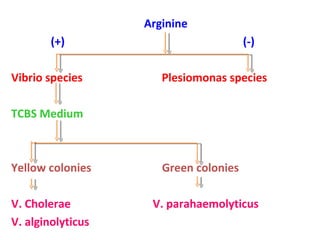
The document discusses identification of pathogenic bacteria in a clinical microbiology laboratory. It provides guidance on identifying common gram positive and gram negative bacteria through microscopic morphology, biochemical tests, and growth characteristics. Key tests discussed include gram stain, catalase, coagulase, optochin sensitivity, bile solubility, and indole for differentiating between staphylococci, streptococci, pneumococci, enterococci, and neisseria.

















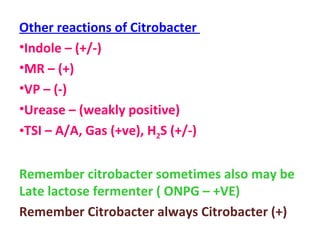

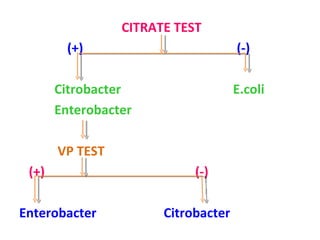
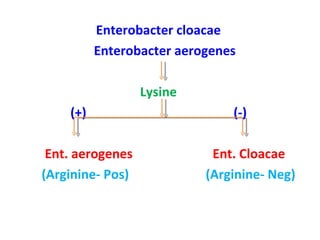

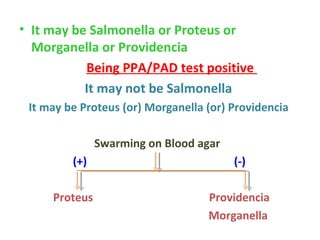








![This presentation was created for those who are working as a laboratory technician in clinical microbiology diagnostics. Also may find useful for UG, PG, DMLT, PGDMLT in Microbiology. Mail ID: [email_address]](https://image.slidesharecdn.com/identificationofbacteria-111011234318-phpapp02/85/Identification-of-bacteria-Bacterial-identification-Lab-identification-of-bacteria-Medical-bacteriology-70-320.jpg)