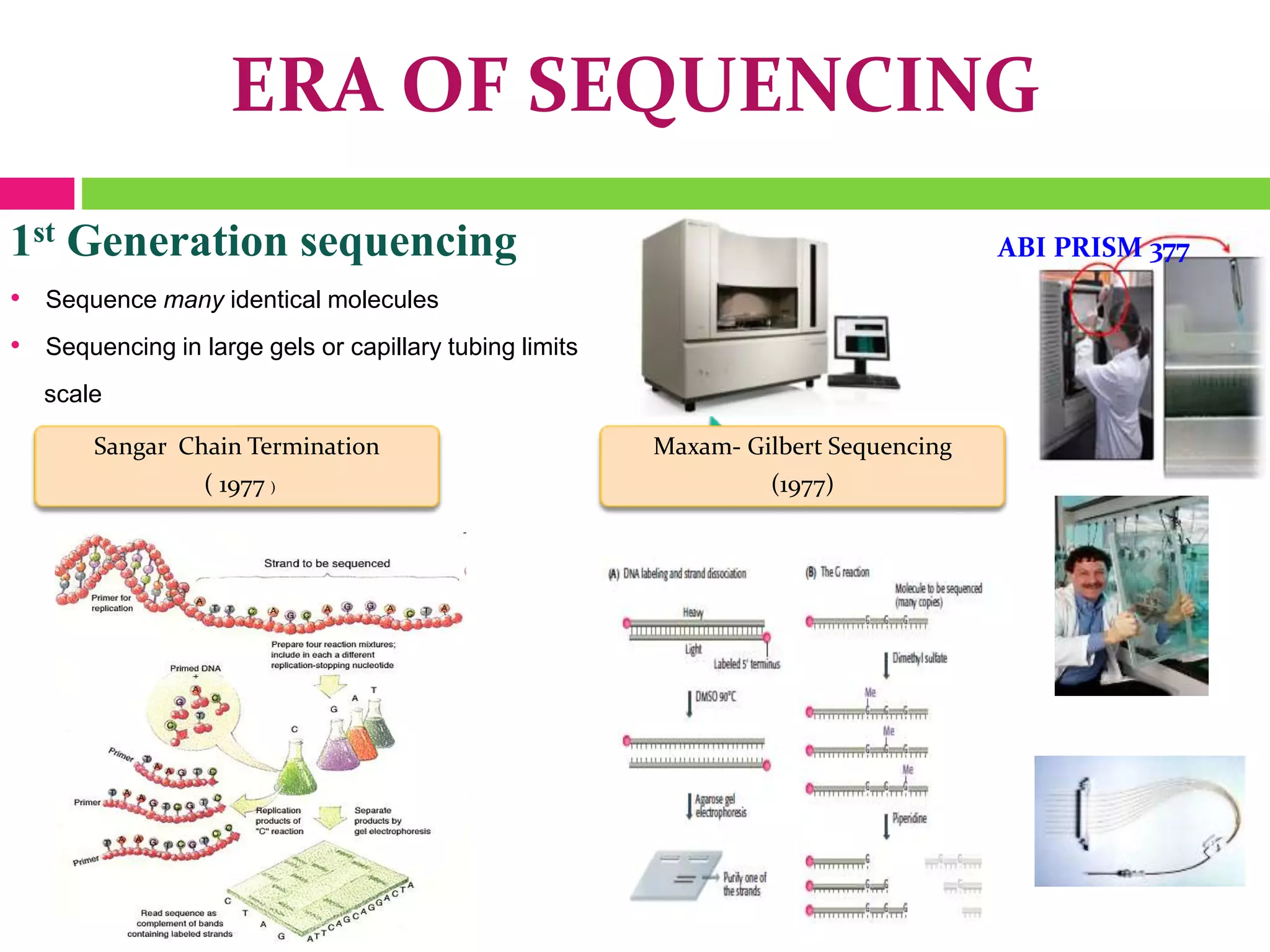
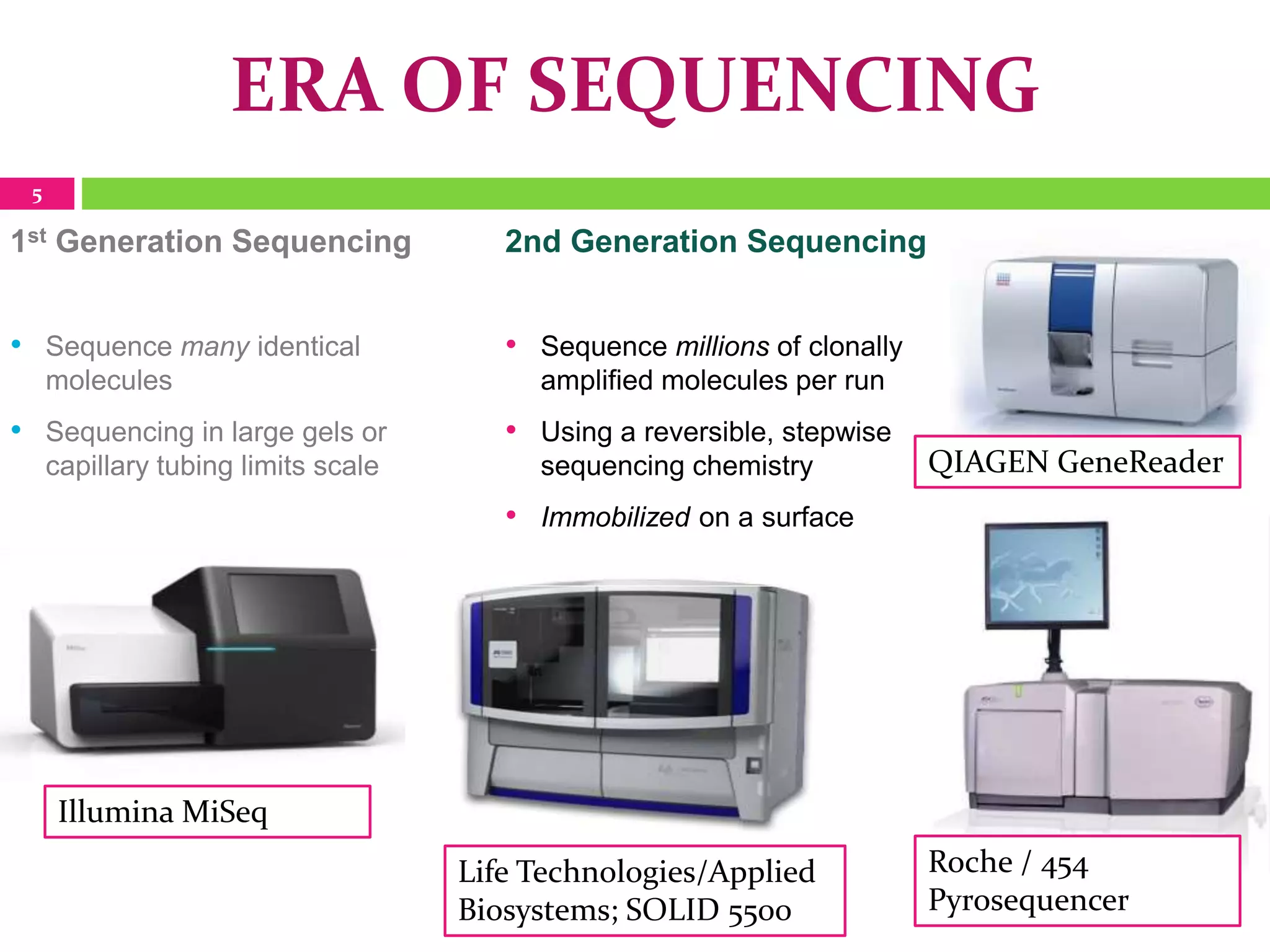
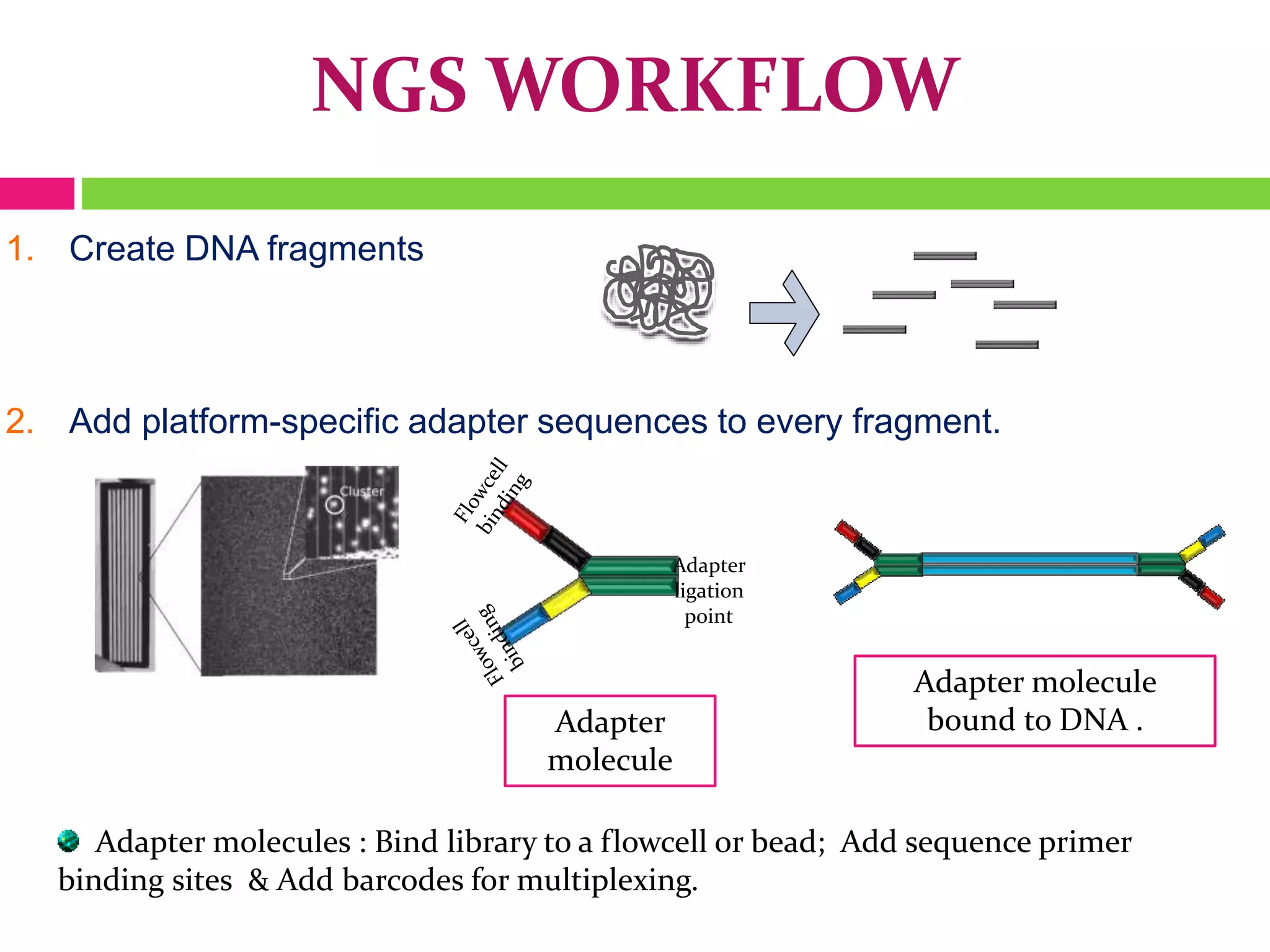
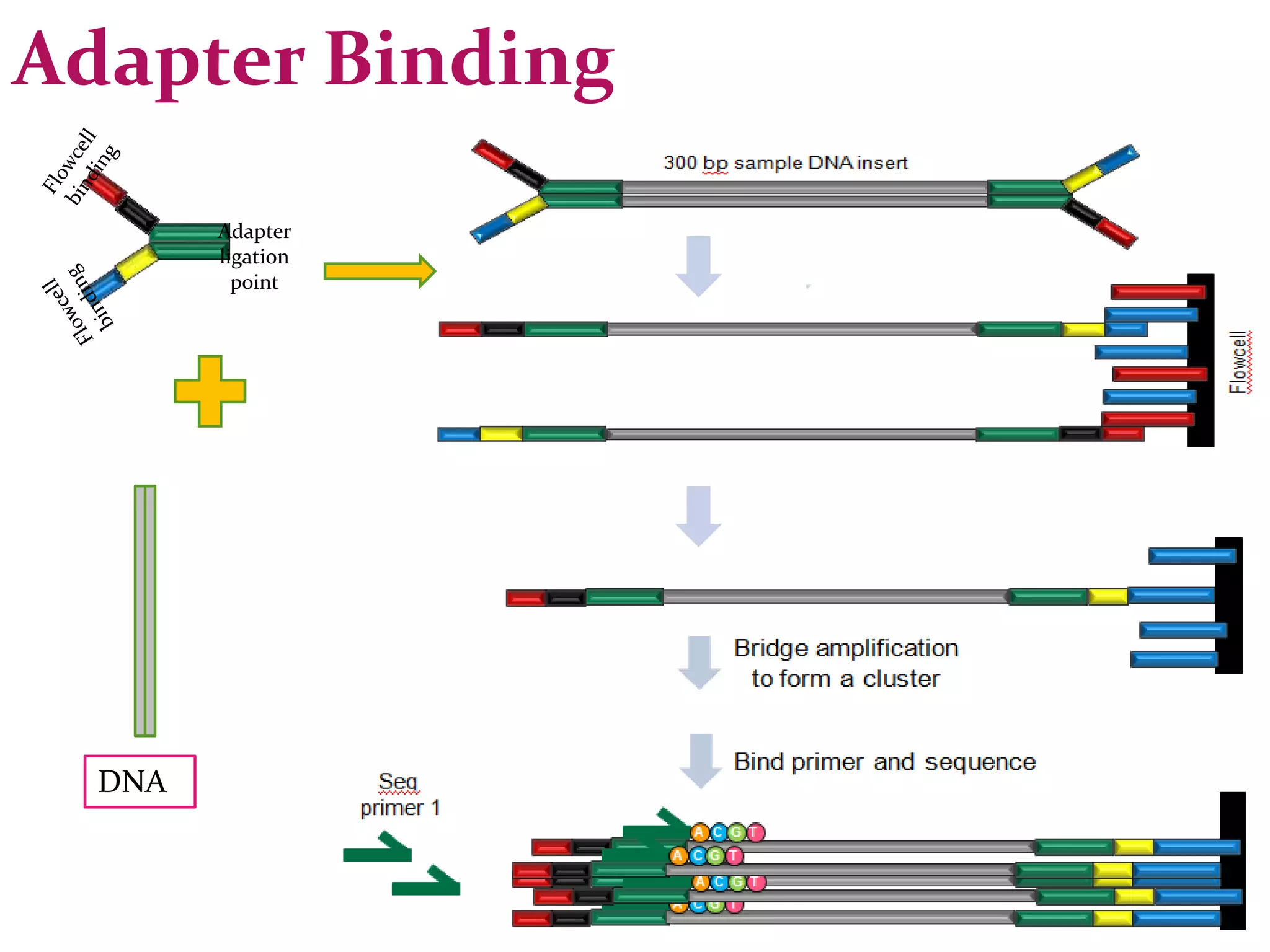
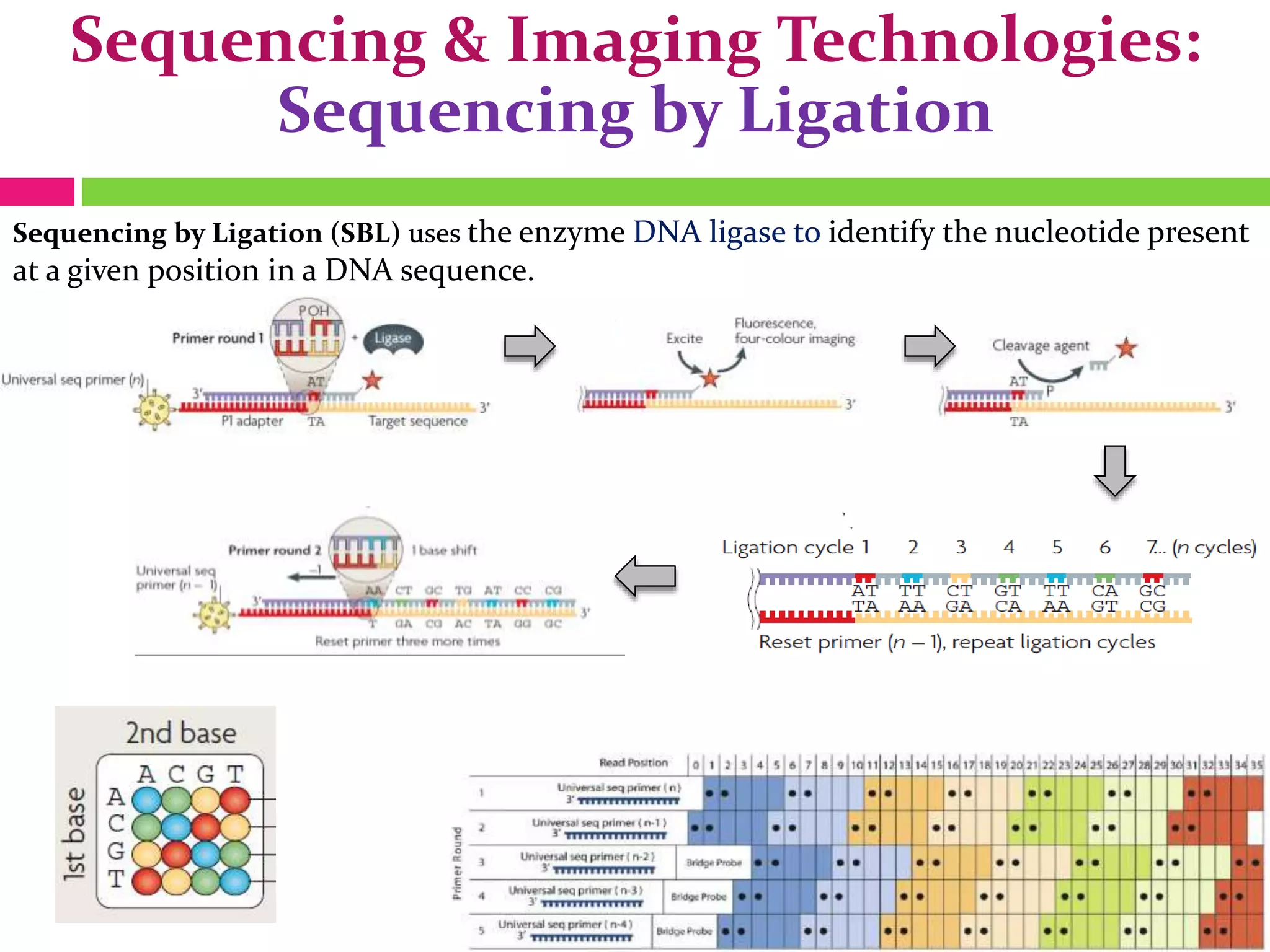
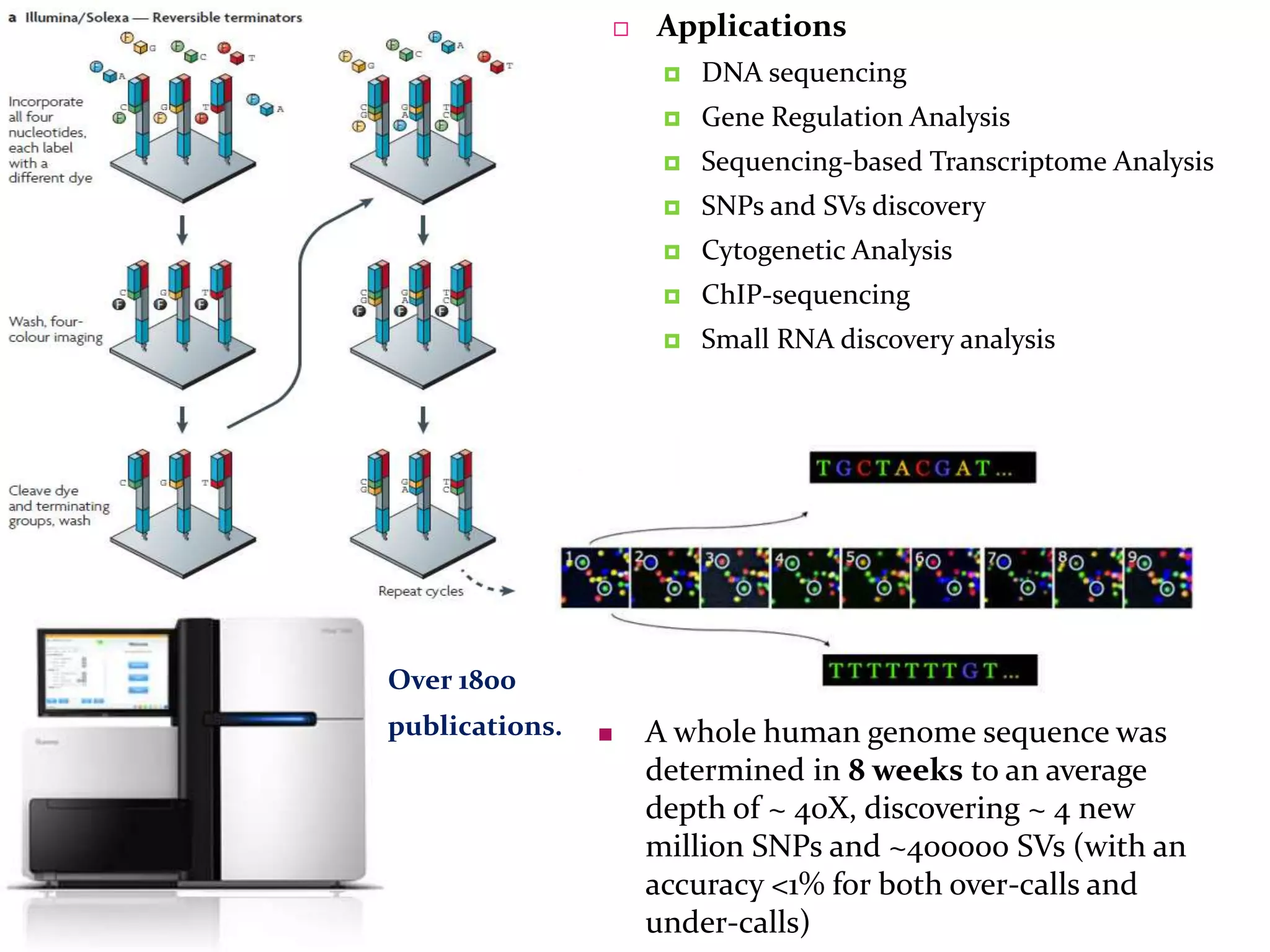
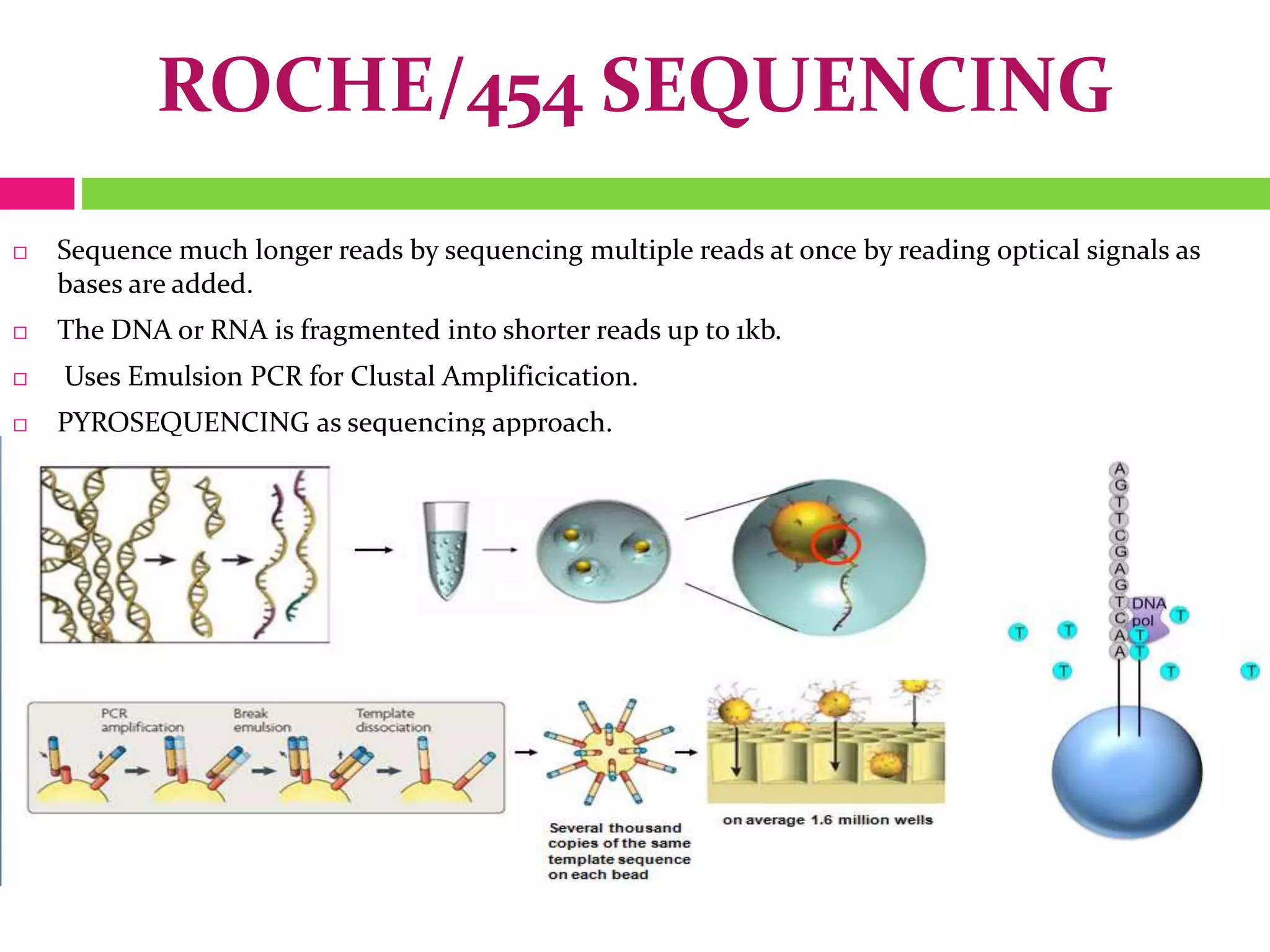
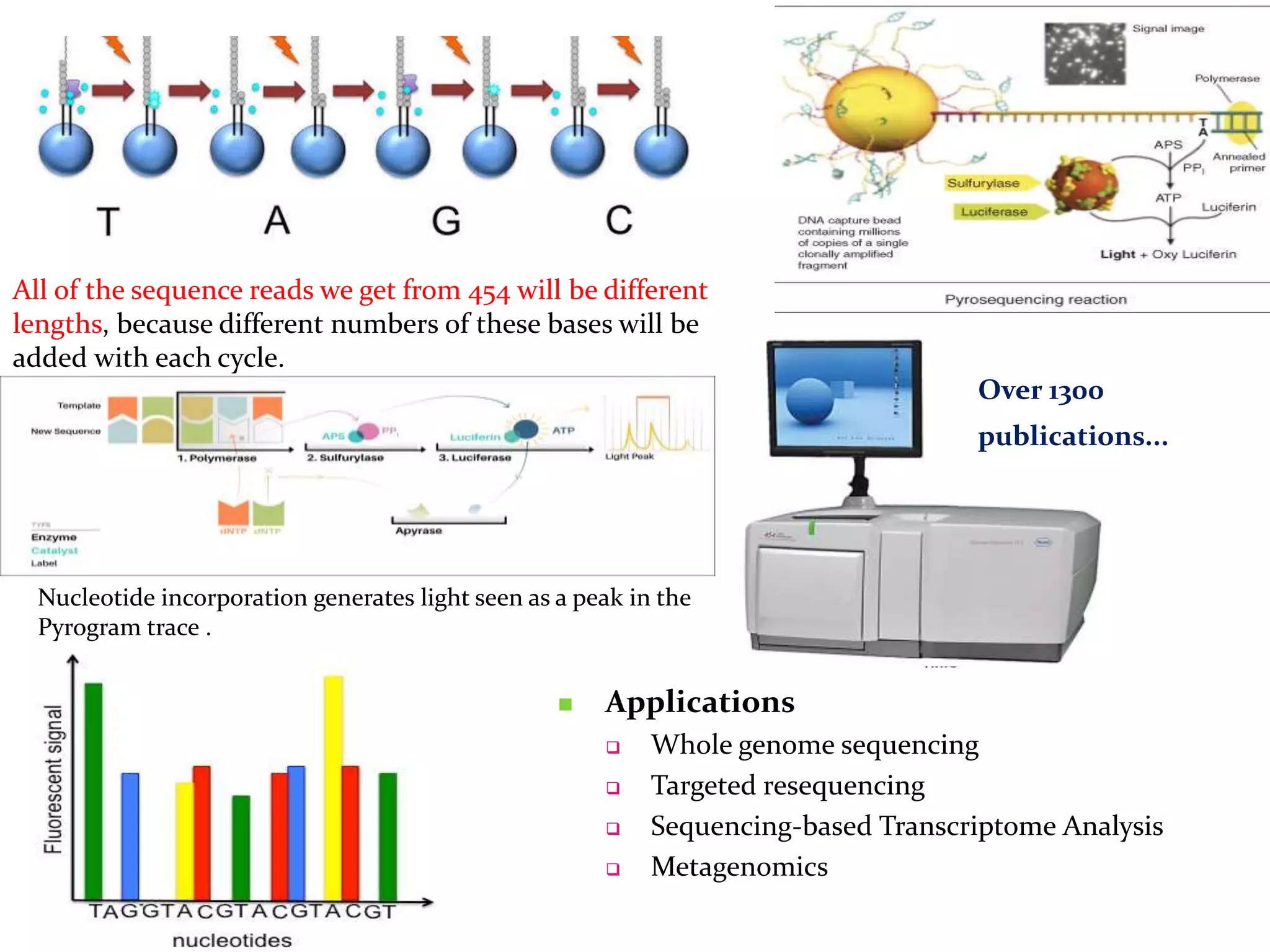
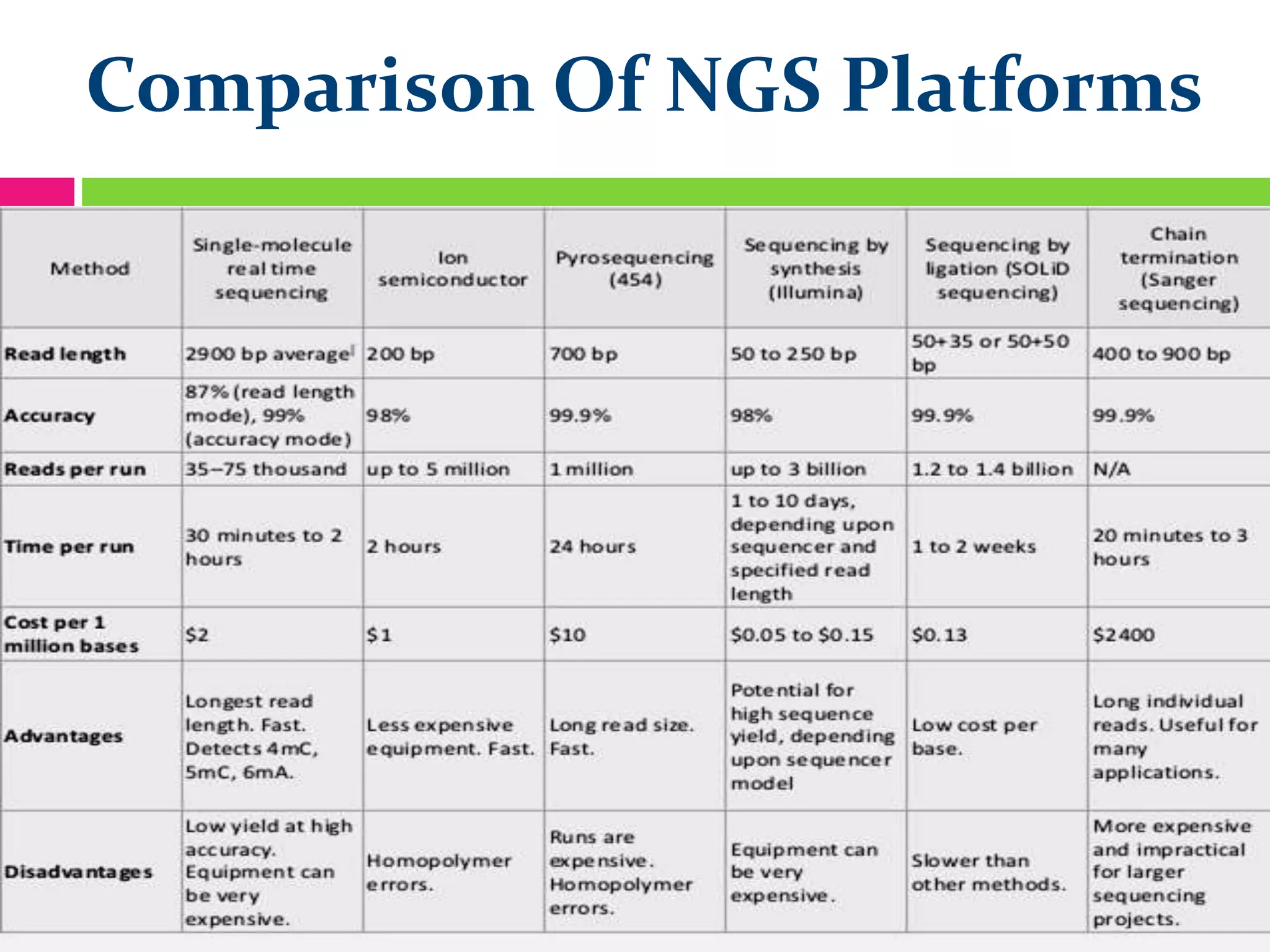
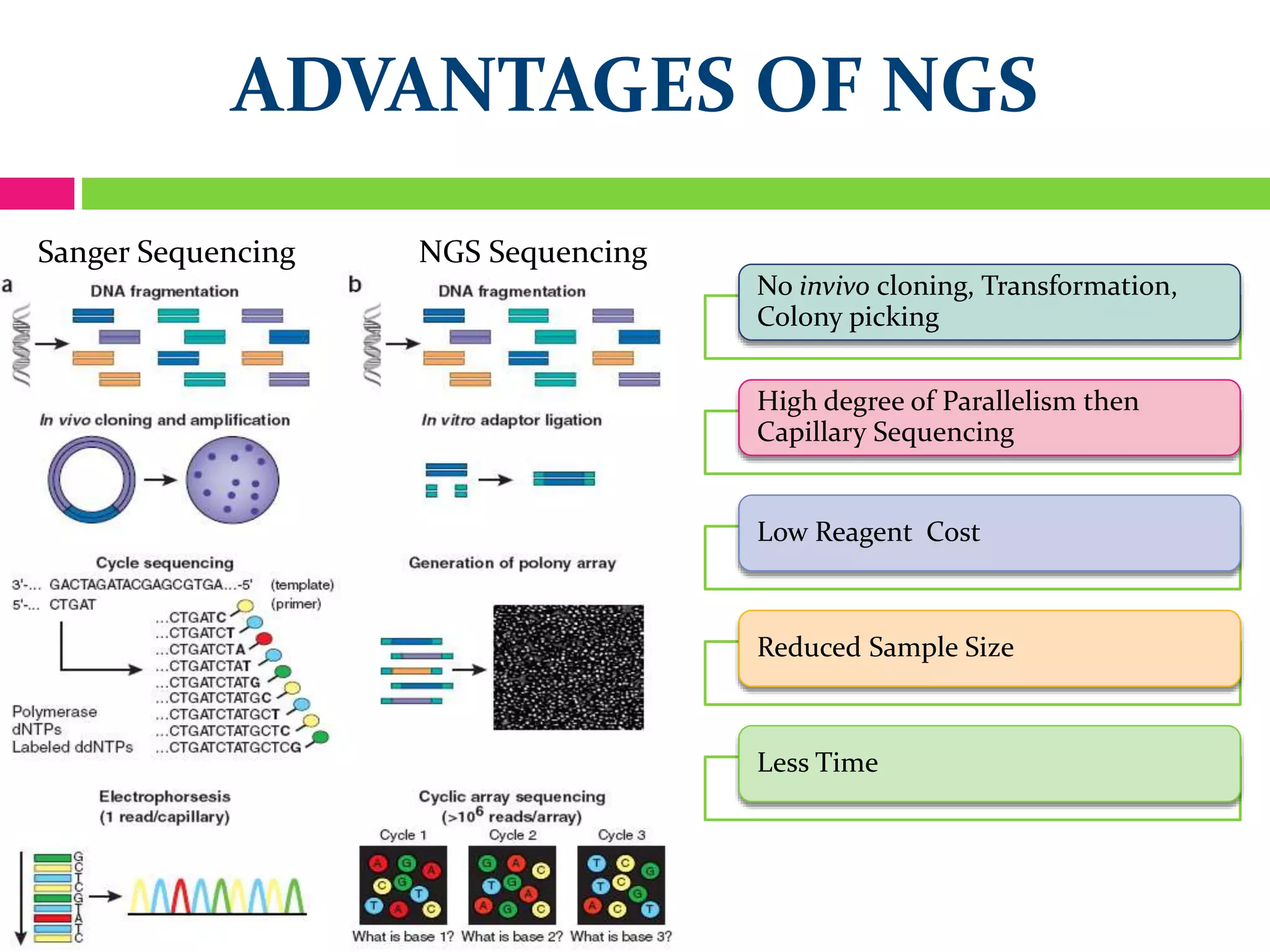
The document provides an overview of DNA sequencing, including the history and advancements from Sanger sequencing to Next Generation Sequencing (NGS) technologies. It outlines various sequencing methods, their workflows, applications, and the advantages of NGS over traditional methods, highlighting its impact on molecular biology and genomics. It also references notable publications and technologies that shaped the evolution of sequencing techniques.