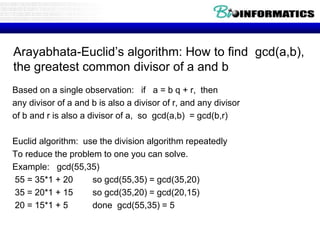
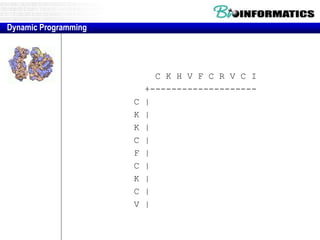
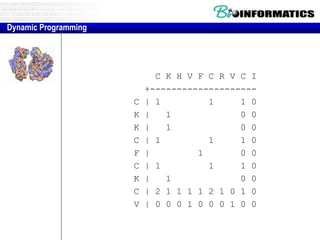
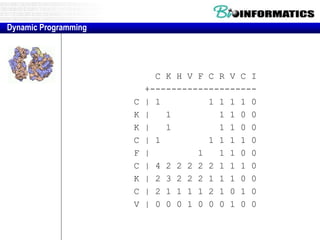
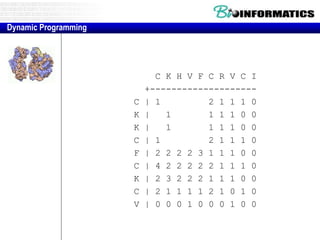
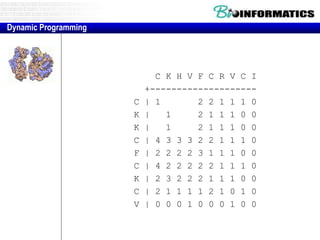
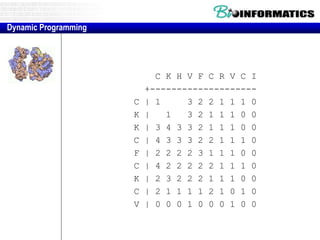
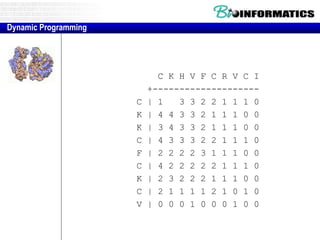
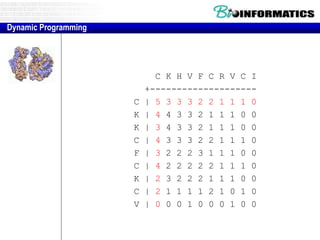
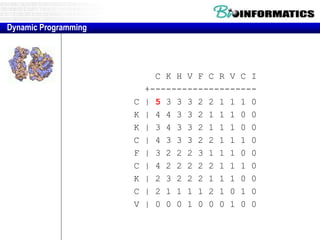
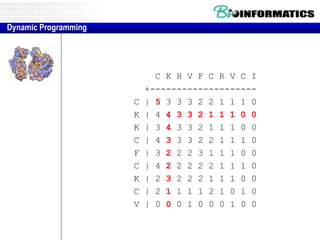
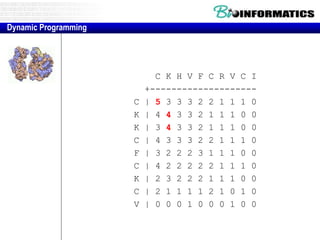
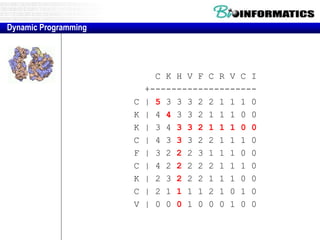
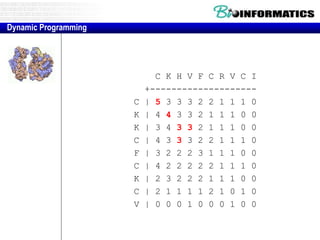
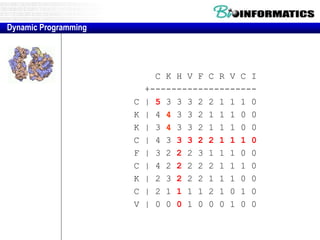
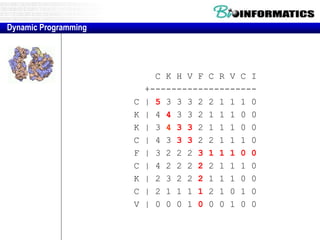
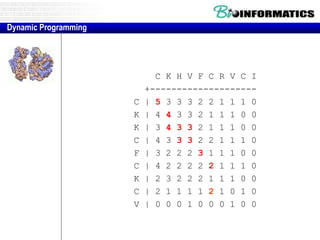
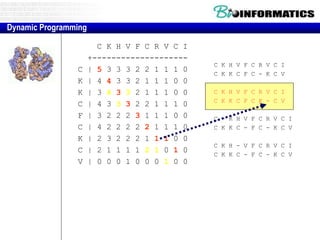
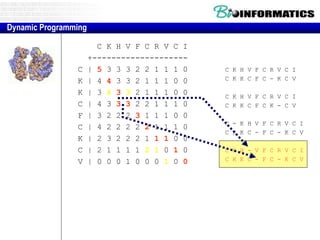
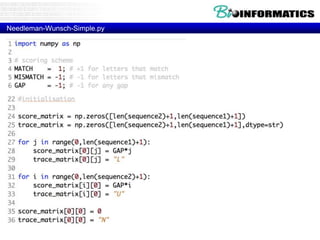
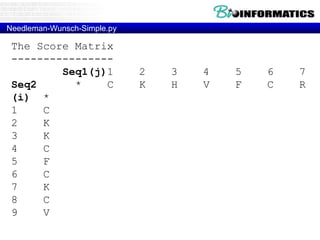
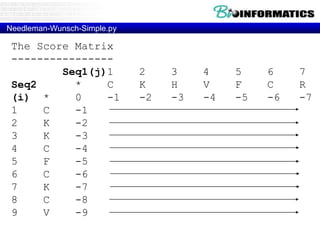
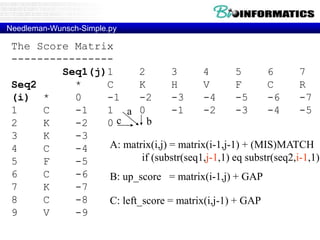
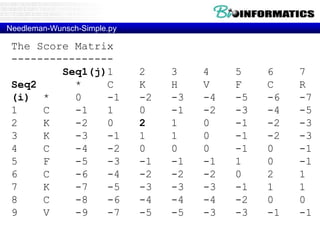
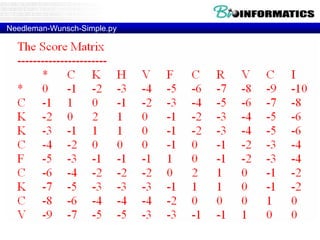
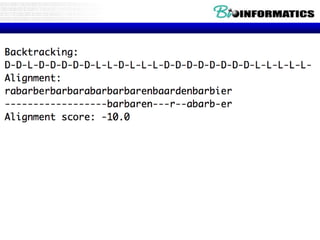
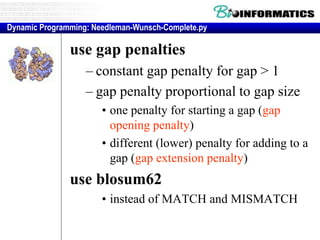
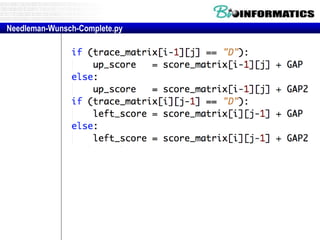
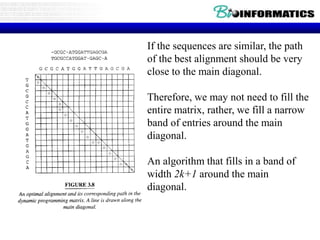
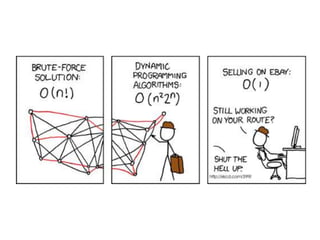
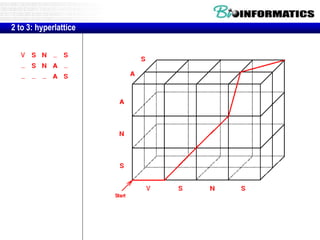
- Dynamic programming is used to find the optimal alignment between two protein sequences by recursively computing sub-alignments and storing them in a lookup table.
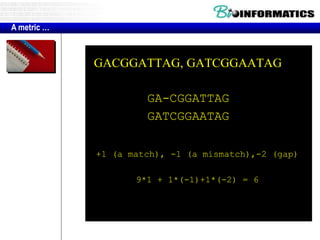
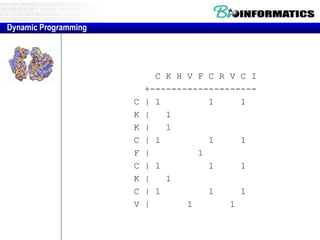
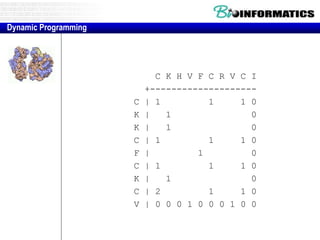
- The example shows calculating the alignment score between a zinc-finger core sequence and a viral sequence fragment by filling a table and tracking the cumulative scores.
- Filling the table from left to right and top to bottom allows reconstructing the highest scoring alignment between the two sequences.









![Bubble Sort Implementation
void BubbleSort(int List[] , int Size) {
int tempInt; // temp variable for swapping list elems
for (int Stop = Size - 1; Stop > 0; Stop--) {
for (int Check = 0; Check < Stop; Check++) { // make a pass
if (List[Check] > List[Check + 1]) { // compare elems
tempInt = List[Check]; // swap if in the
List[Check] = List[Check + 1]; // wrong order
List[Check + 1] = tempInt;
}
}
}
}
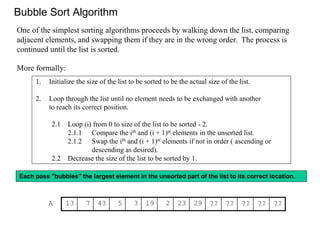
Bubblesort compares and swaps adjacent elements; simple but not very efficient.
Efficiency note: the outer loop could be modified to exit if the list is already sorted.
Here is an ascending-order implementation of the bubblesort algorithm for integer arrays:](https://image.slidesharecdn.com/2016bioinformaticsialignmentswimvancriekinge-161024084206/85/2016-bioinformatics-i_alignments_wim_vancriekinge-19-320.jpg)




























![****** MULTIPLE ALIGNMENT MENU ******
1. Do complete multiple alignment now (Slow/Accurate)
2. Produce guide tree file only
3. Do alignment using old guide tree file
4. Toggle Slow/Fast pairwise alignments = SLOW
5. Pairwise alignment parameters
6. Multiple alignment parameters
7. Reset gaps between alignments? = OFF
8. Toggle screen display = ON
9. Output format options
S. Execute a system command
H. HELP
or press [RETURN] to go back to main menu
Your choice:
Running ClustalW](https://image.slidesharecdn.com/2016bioinformaticsialignmentswimvancriekinge-161024084206/85/2016-bioinformatics-i_alignments_wim_vancriekinge-102-320.jpg)


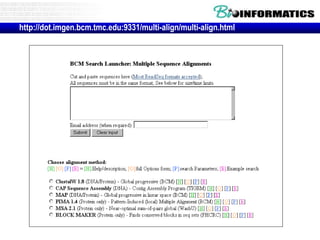











![• Go to http://www.ncbi.nlm.nih.gov/BLAST
• Choose BLAST 2 sequences
• In the program,
[1] choose blastp (protein search) or blastn (for DNA)
[2] paste in your accession numbers
(or use FASTA format)
[3] select optional parameters, such as
--BLOSU62 matrix is default for proteins
try PAM250 for distantly related proteins
--gap creation and extension penalties
[4] click “align”
Practical guide to pairwise alignment:
the “BLAST 2 sequences” website](https://image.slidesharecdn.com/2016bioinformaticsialignmentswimvancriekinge-161024084206/85/2016-bioinformatics-i_alignments_wim_vancriekinge-117-320.jpg)