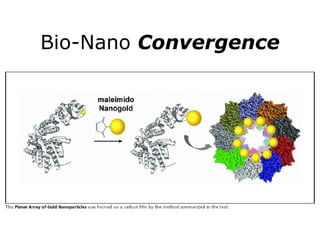
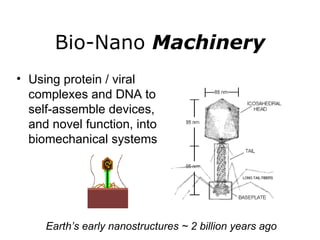
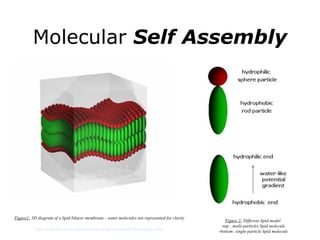
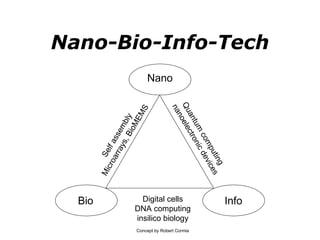
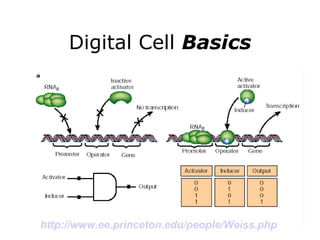
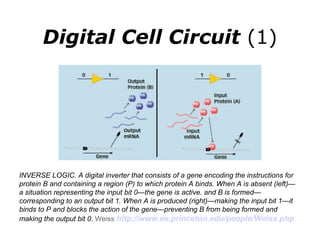
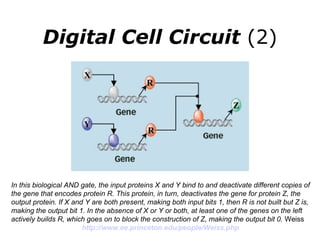
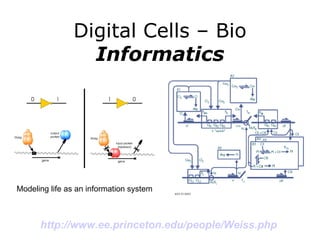
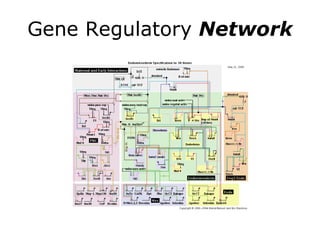
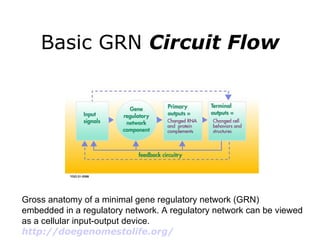
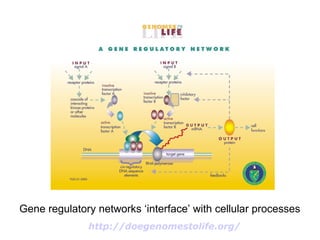
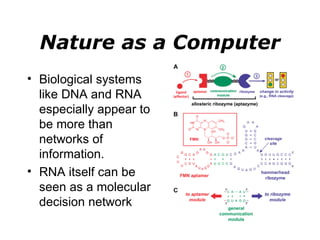
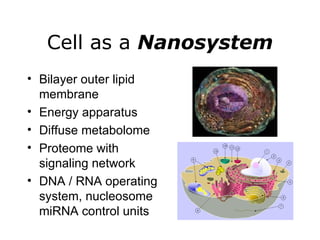
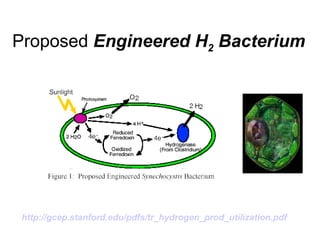
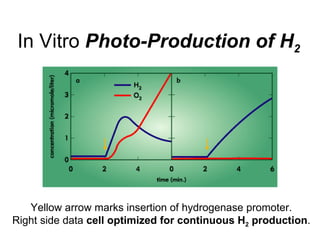
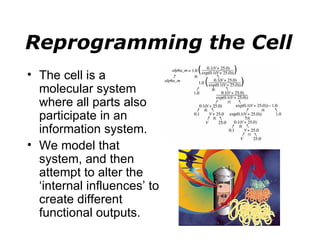
Synthetic biology builds on nanotechnology and biotechnology by adding information technology to model and modify biological systems at the genetic level. It aims to program cells by reengineering genomes and integrating biology with nanotechnology. Researchers can model gene networks, validate circuits, and alter genes to design new cellular functions. The next frontier is bringing such innovations to higher organisms using stem cells. The overall goal is to understand and reprogram biology as an information processing system at the molecular scale.