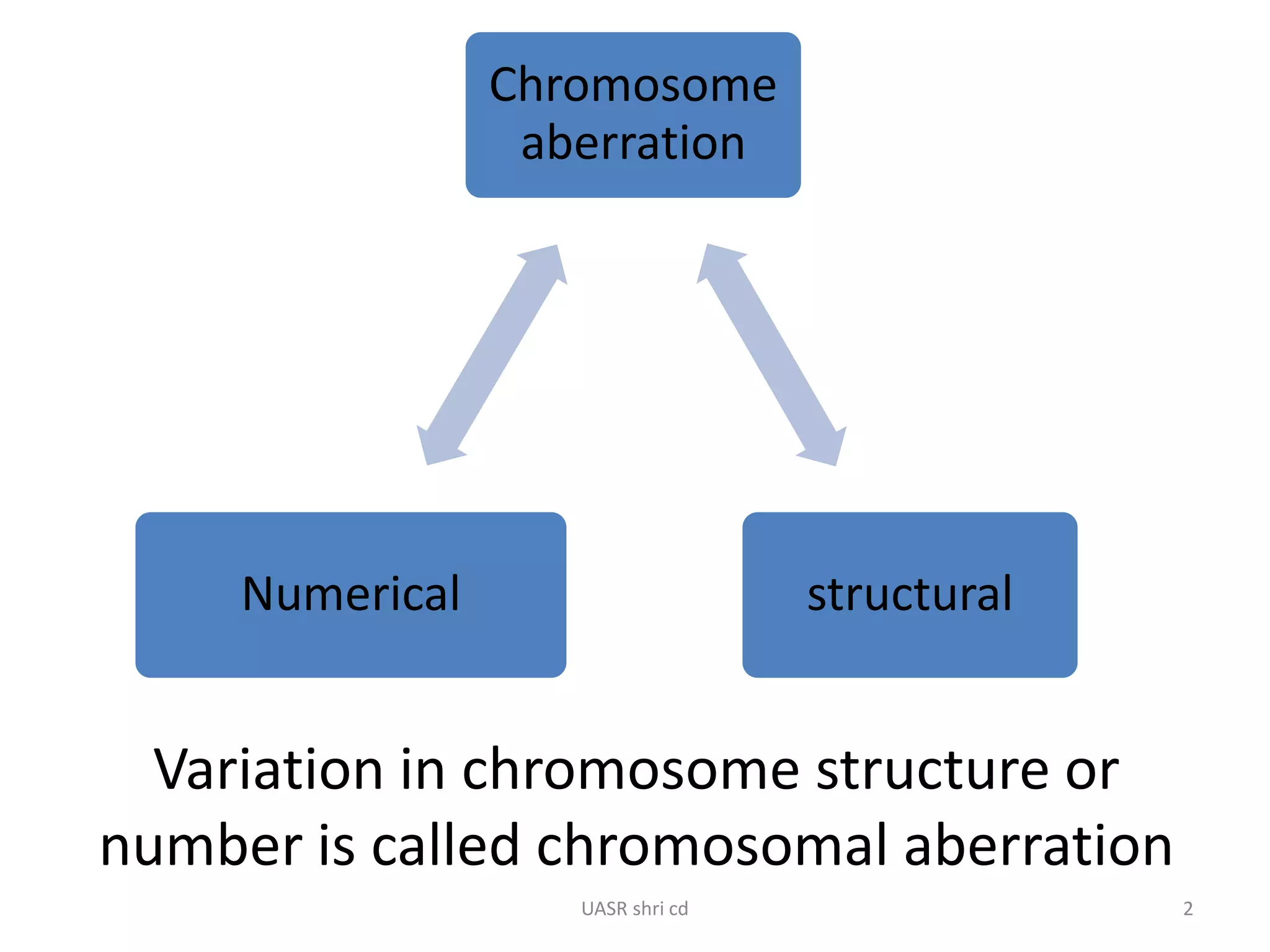
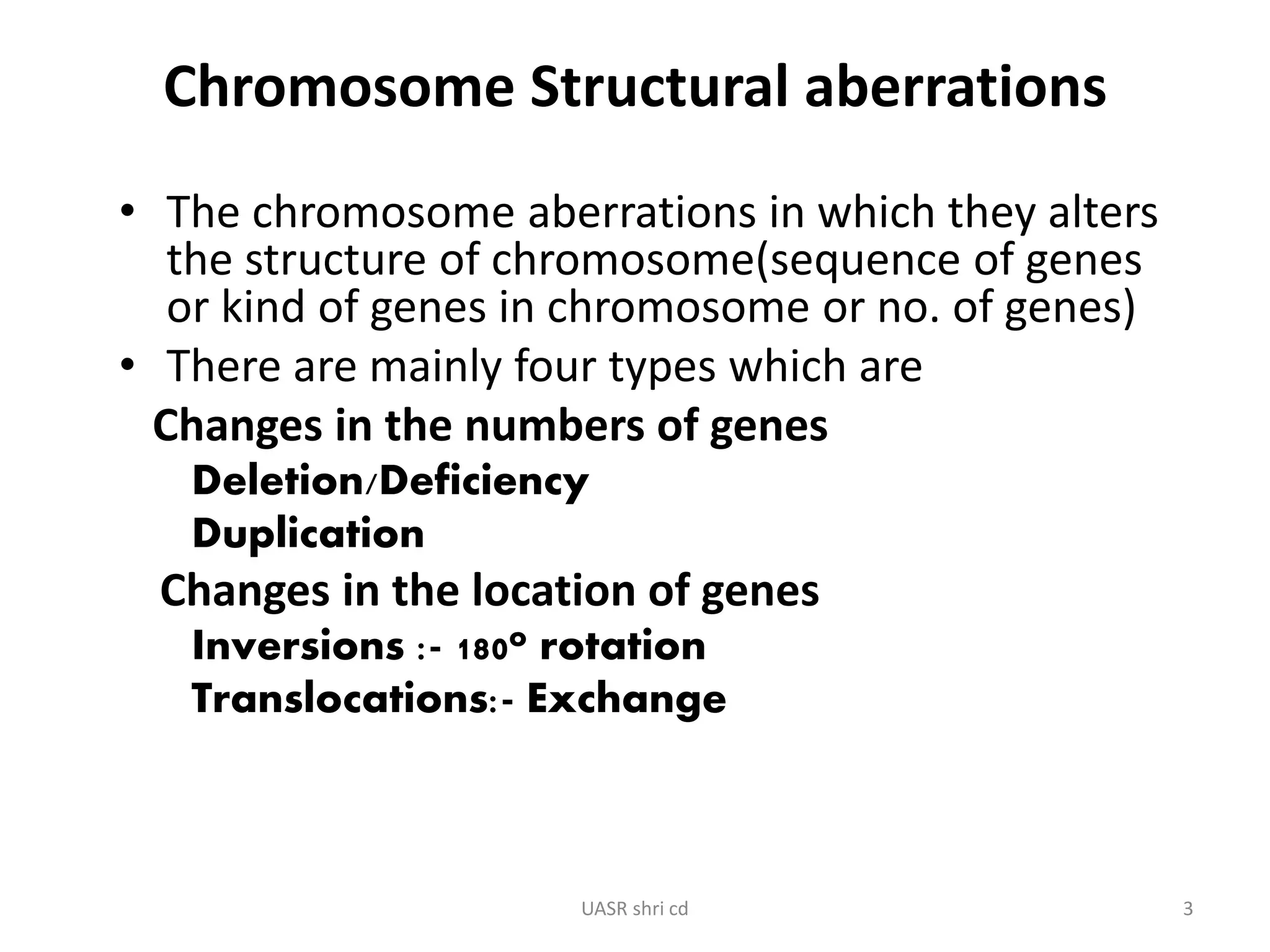
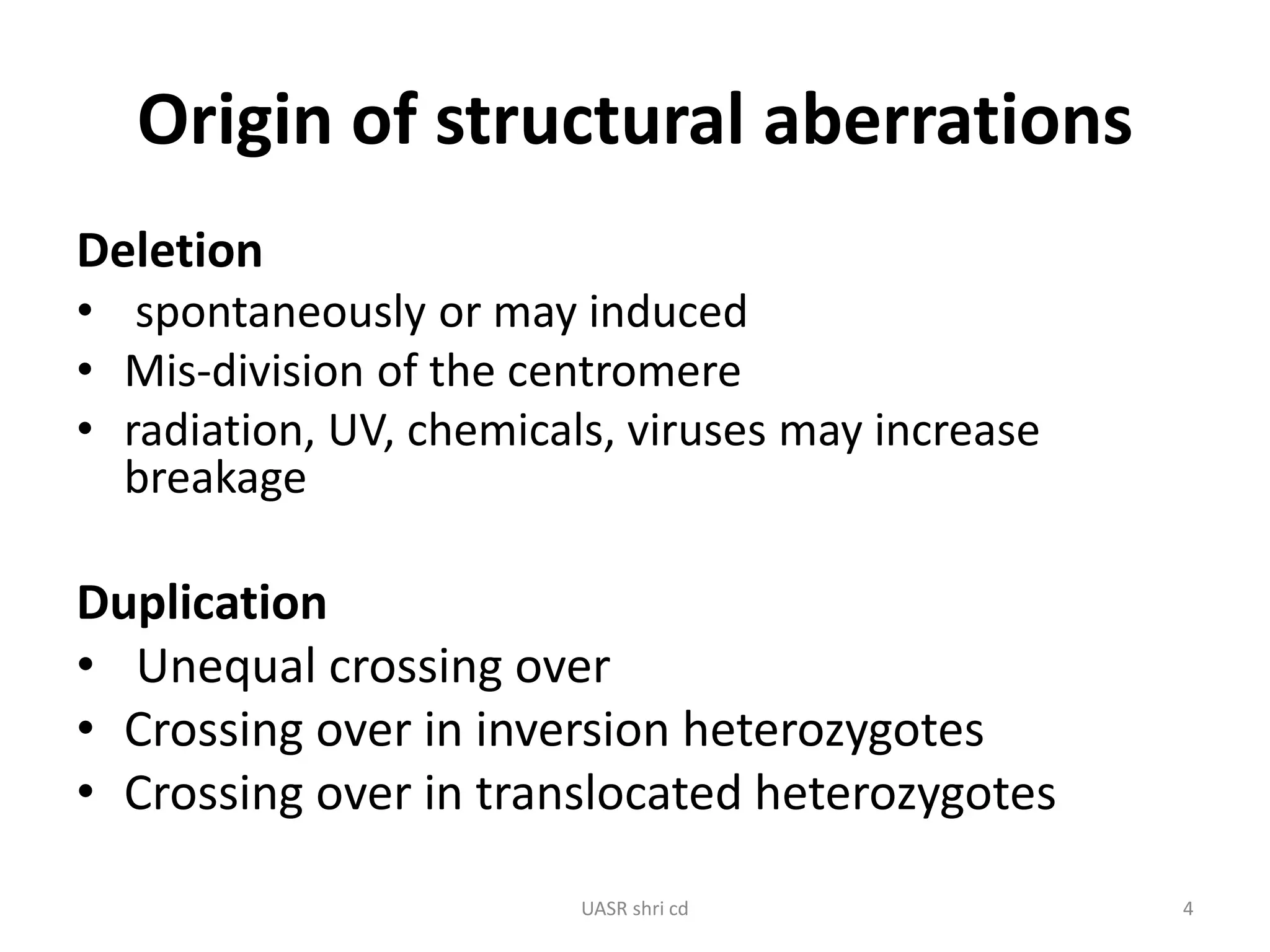
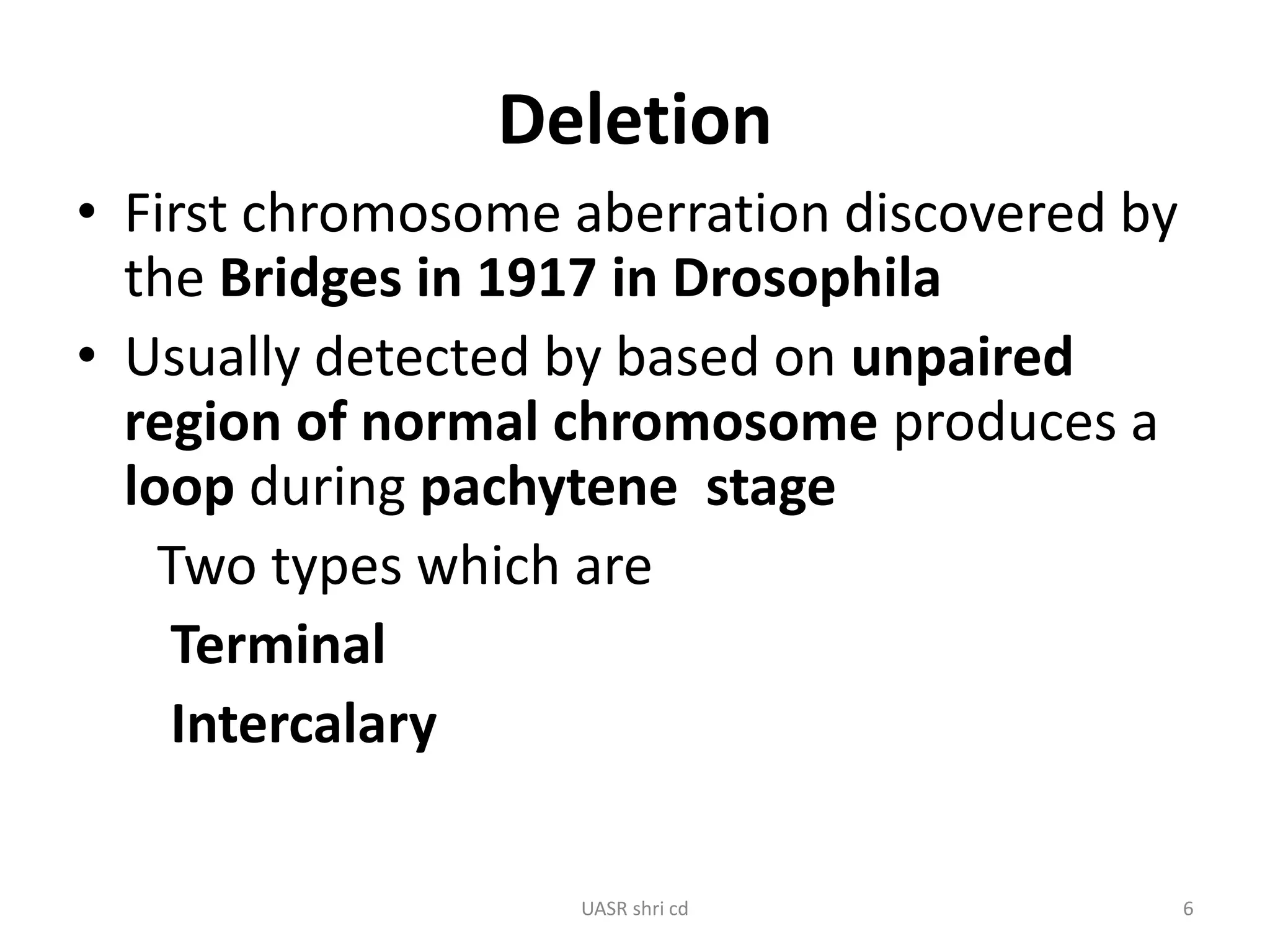
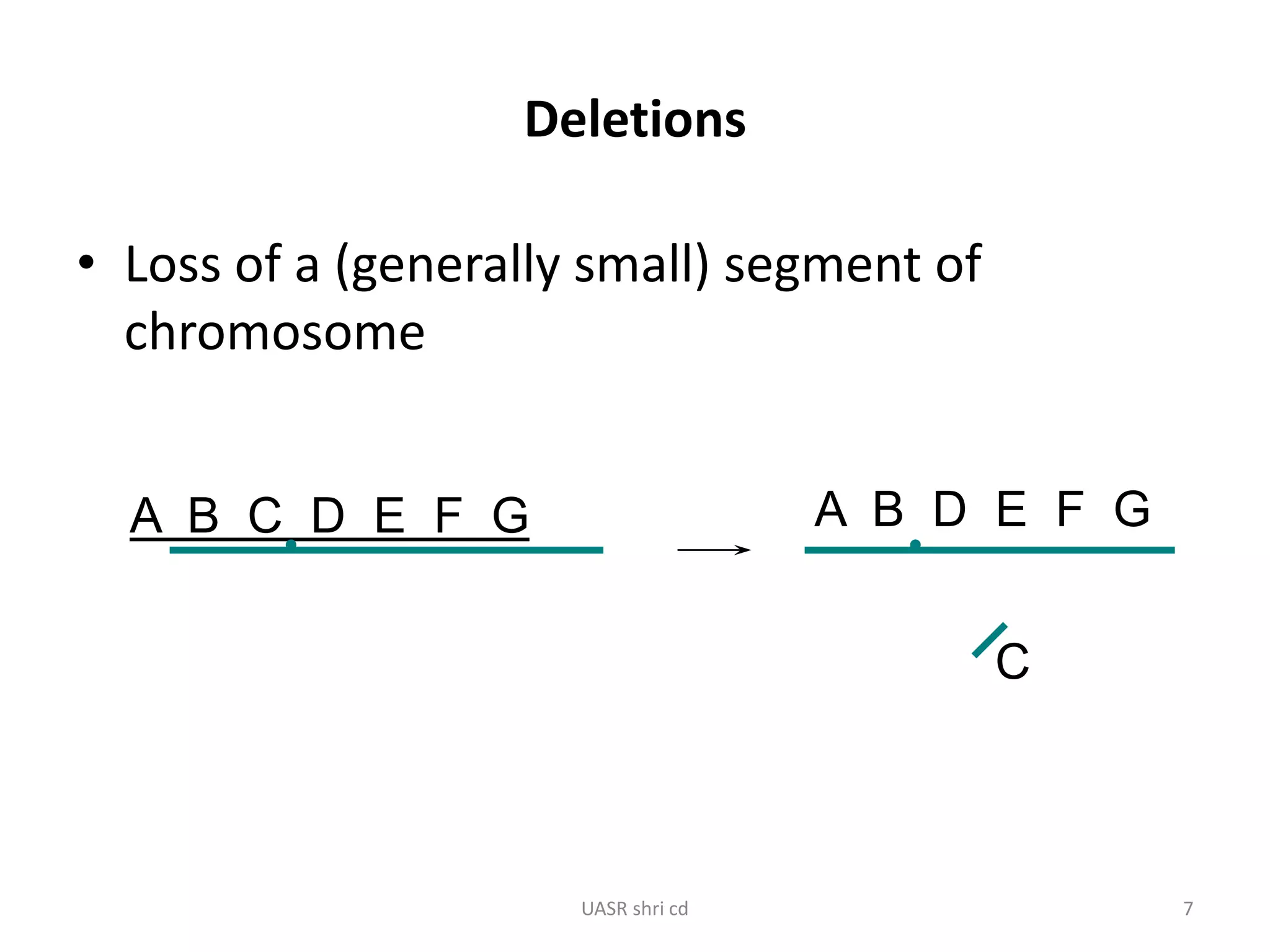
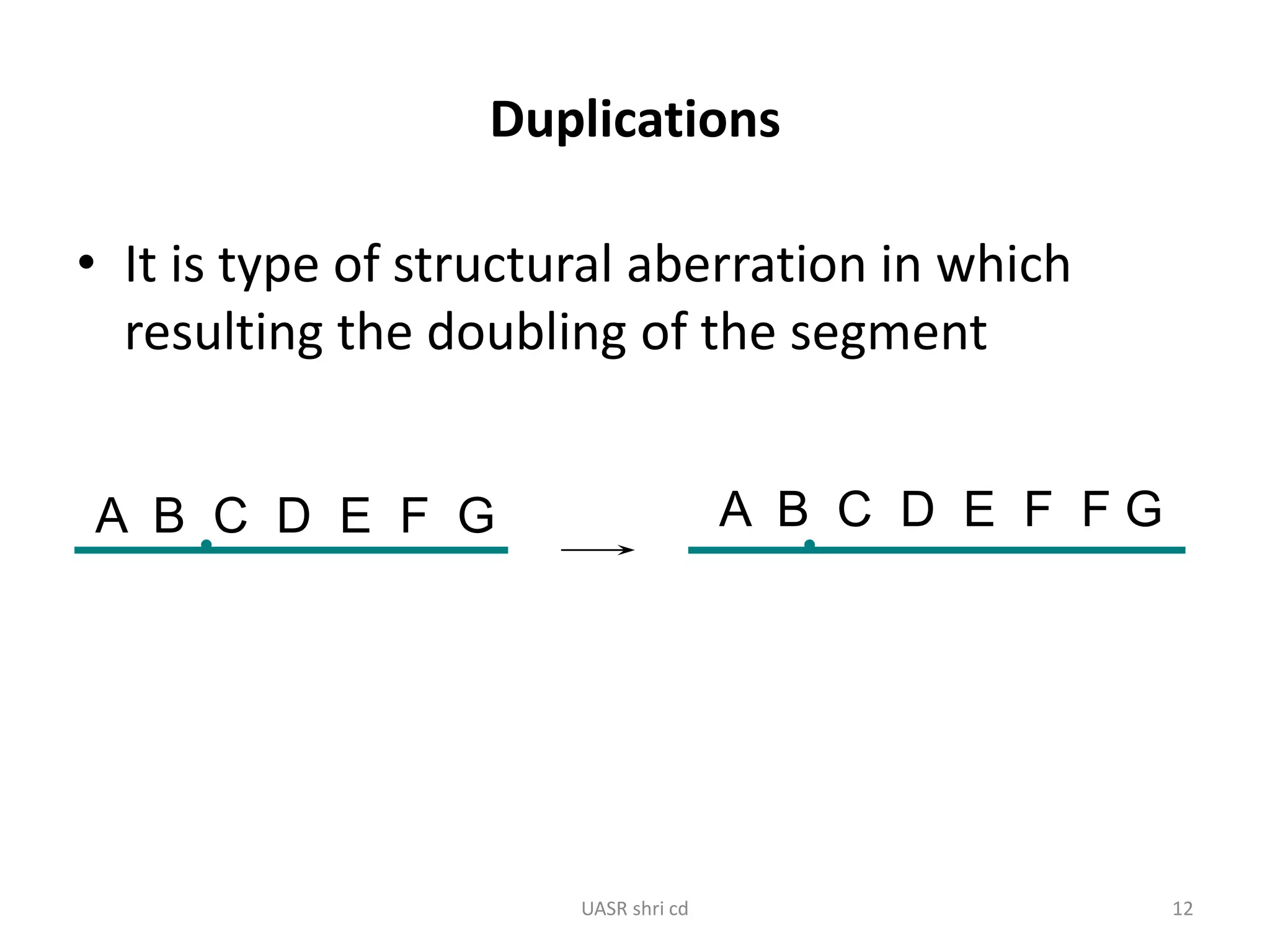
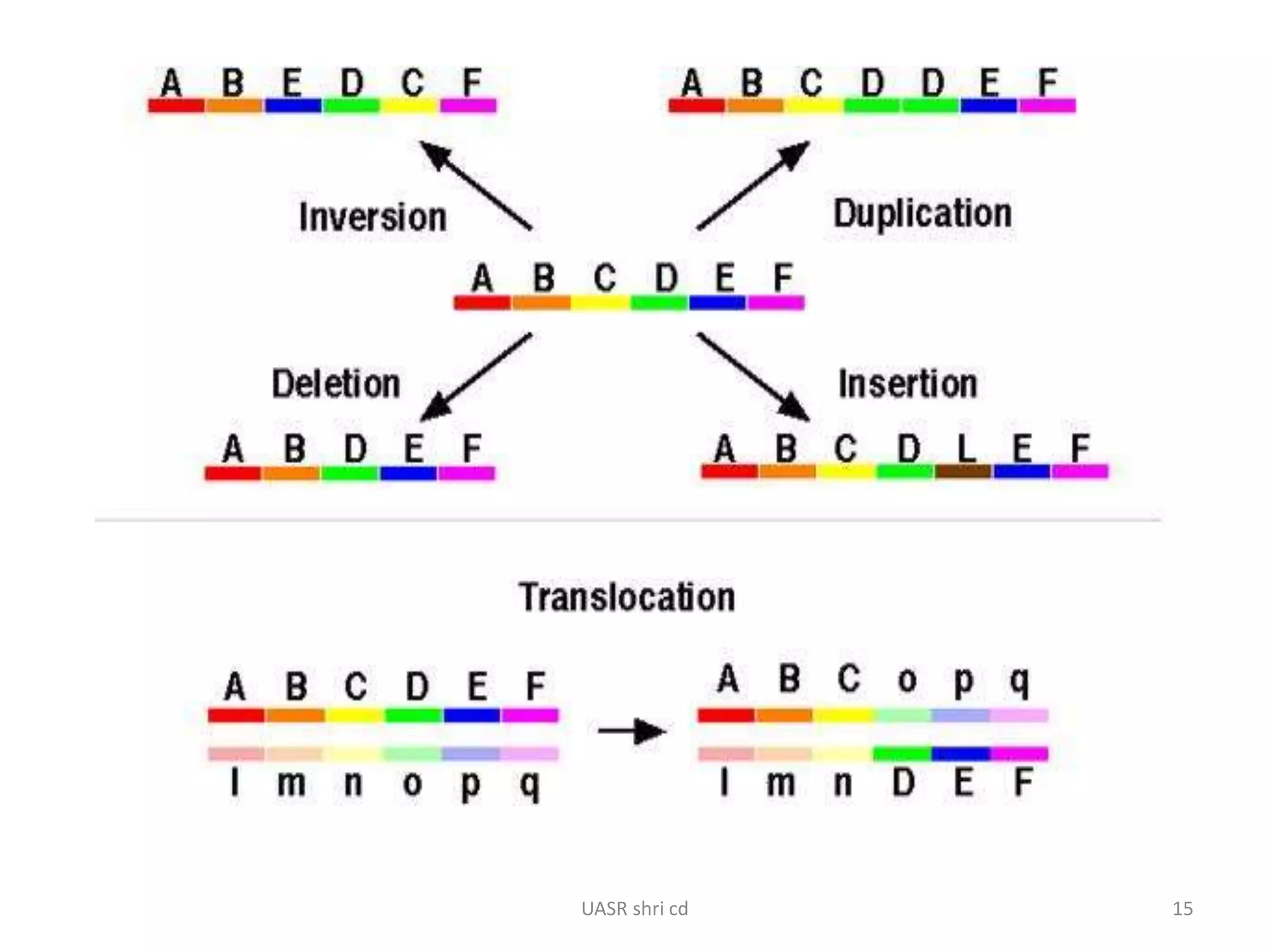
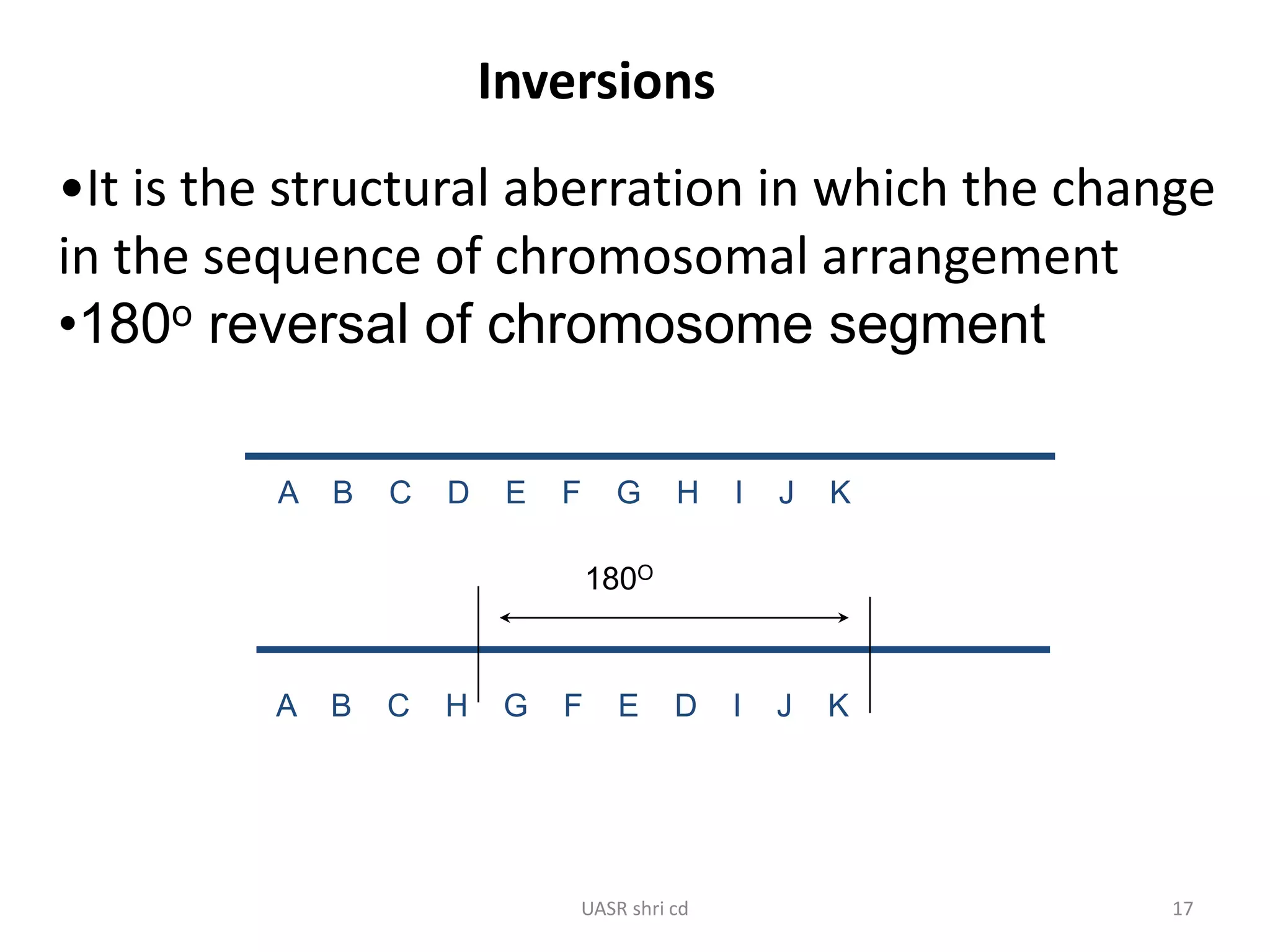
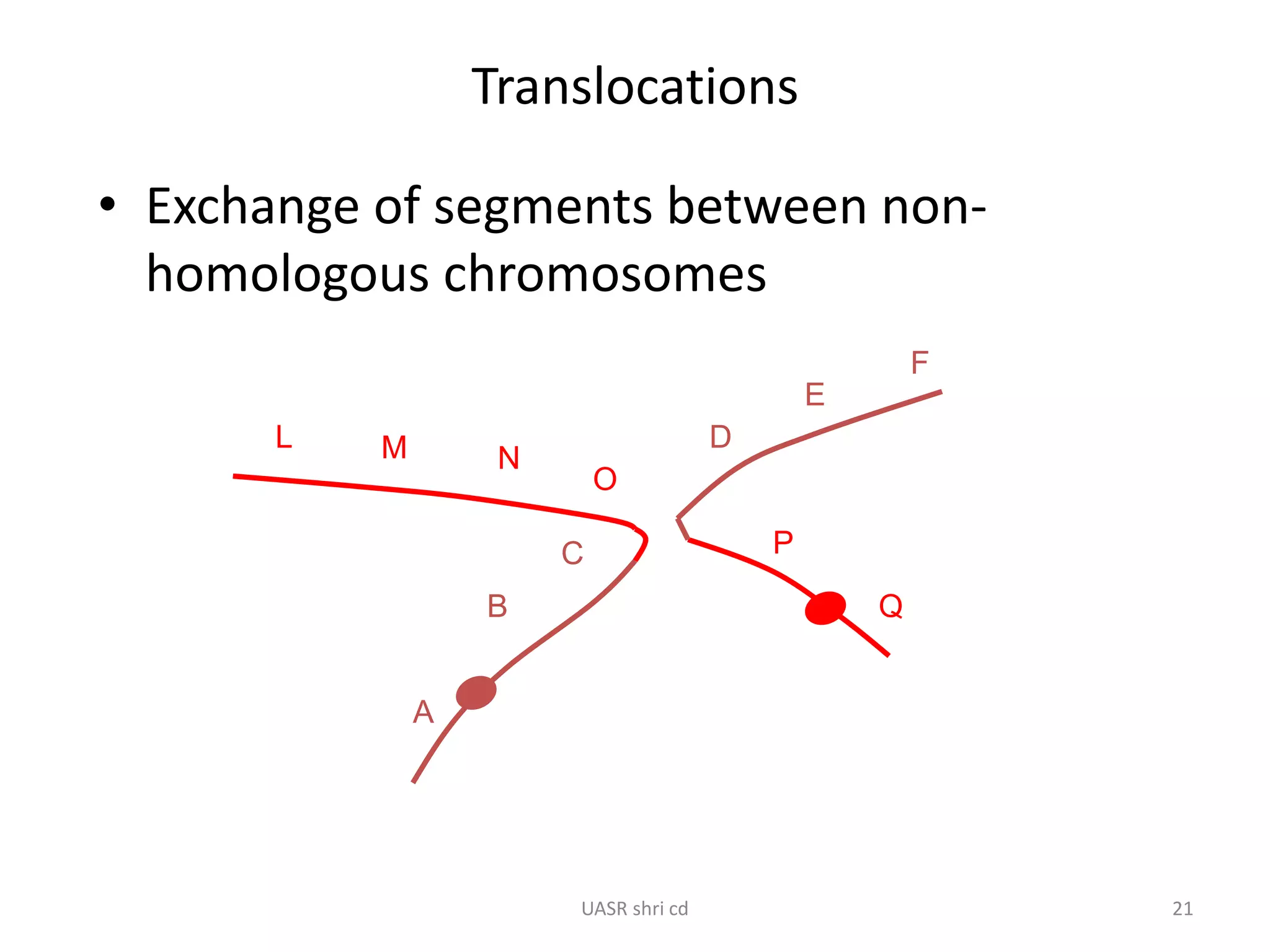
Structural chromosomal aberrations and their role in plant breeding was presented. There are four main types of structural aberrations: changes in gene number, deletions, duplications, and changes in gene location through inversions and translocations. These aberrations can occur spontaneously or be induced, and can provide benefits in plant breeding such as increasing desirable gene dosage or producing new genes. However, they can also lead to harmful effects like male sterility or recessive lethals in diploid organisms.