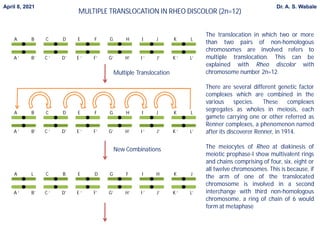
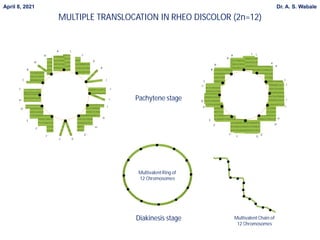
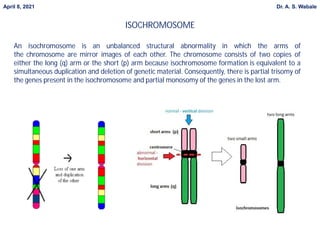
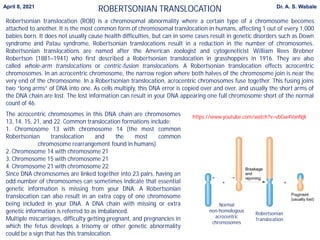
Here are the key points to cover in the assignment:
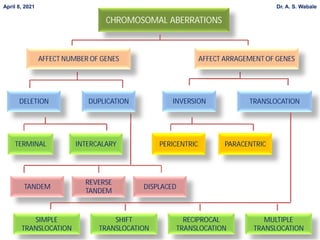
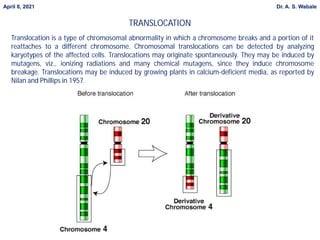
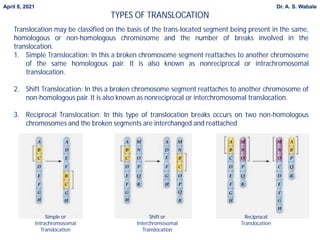
1. Briefly explain the different types of structural changes that can occur in chromosomes - deletion, duplication, inversion, translocation.
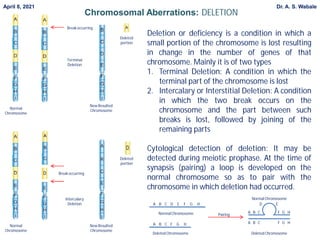
2. Explain deletion in more detail - its types (terminal, intercalary), cytological detection using looping during meiosis, and genetic effects of changing gene number.
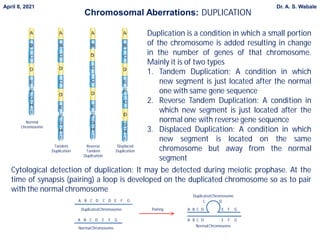
3. Explain duplication in more detail - its types (tandem, reverse, displaced), cytological detection using looping, and effects of changing gene number.
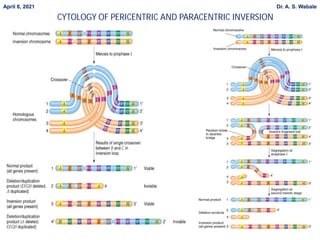
4. Explain inversion in more detail - its types based on centromere involvement (pericentric, paracentric), and that it does not change overall genetic material.
5. Provide detailed explanation