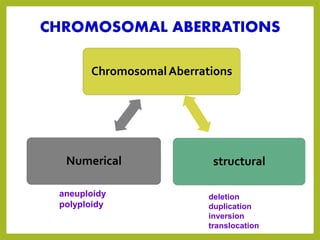
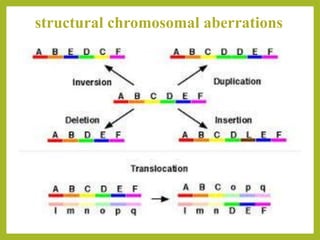
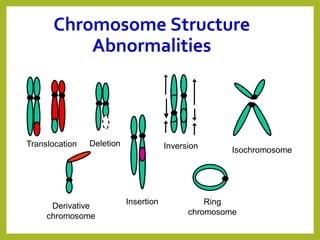
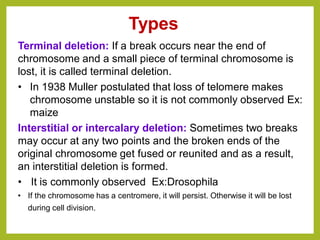
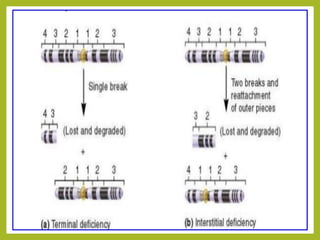
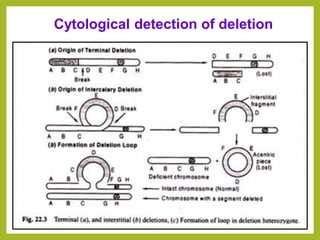
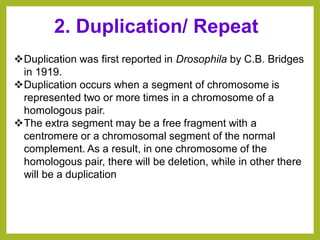
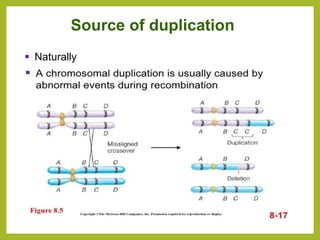
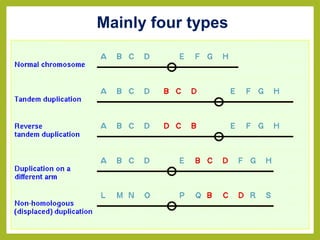
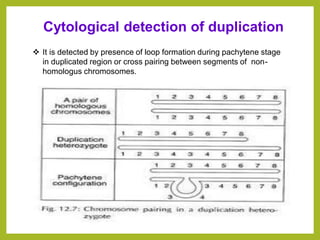
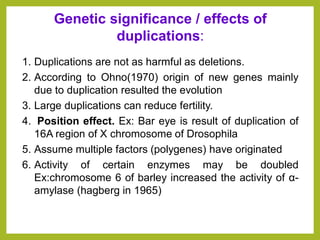
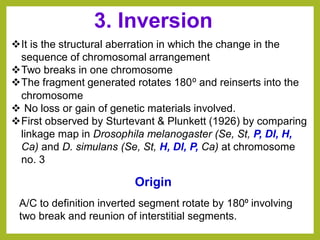
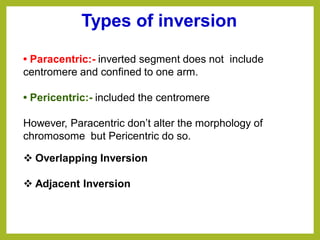
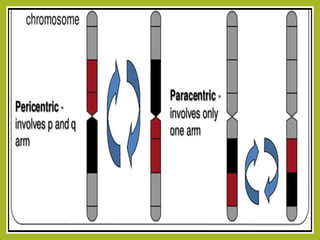
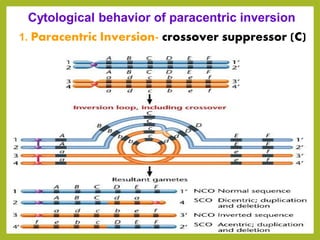
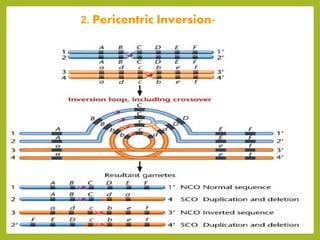
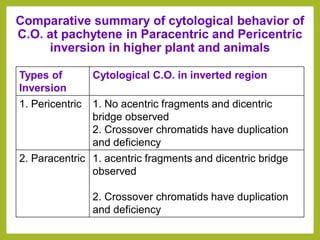
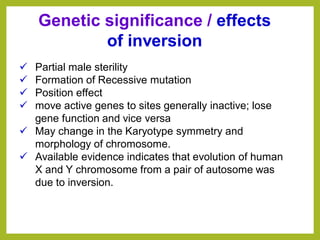
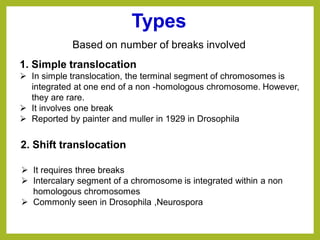
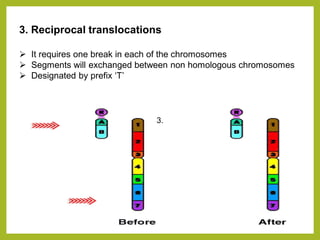
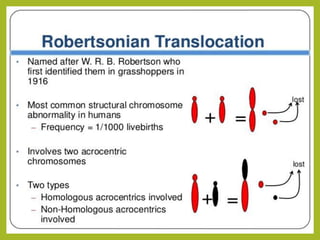
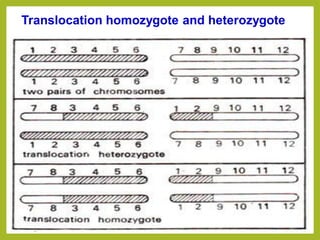
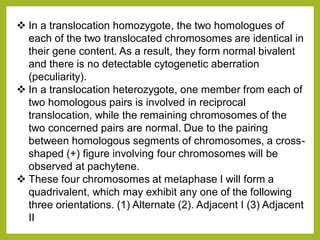
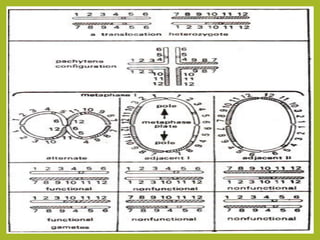
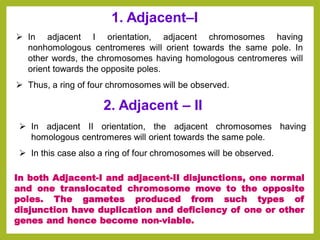
Chromosomal aberrations refer to changes in chromosome structure or number that can result in genetic disorders. The document discusses several types of chromosomal aberrations including numerical abnormalities like aneuploidy and structural abnormalities like deletions, duplications, inversions, and translocations. Deletions occur when a segment of a chromosome is lost, duplications when a segment is repeated, inversions when a segment rotates 180 degrees, and translocations when a segment is transferred to a non-homologous chromosome. These aberrations can impact fertility, gene expression, and viability and have been implicated in genetic disorders and evolution. The document provides detailed explanations and examples of each type of aberration.