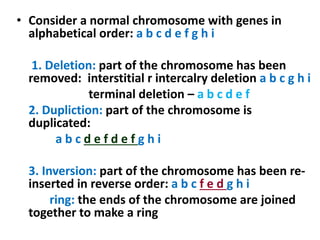
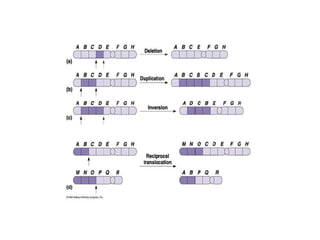
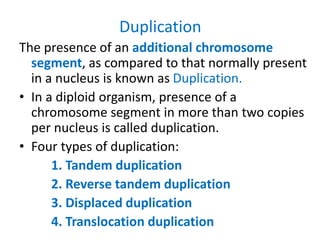
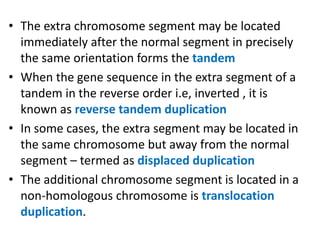
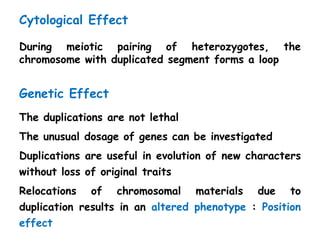
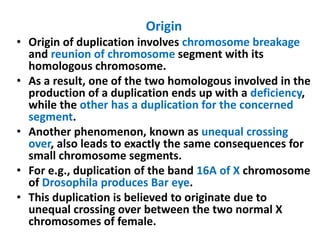
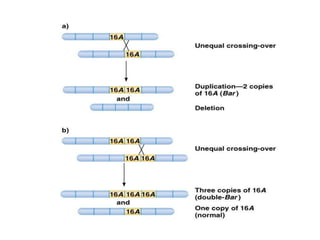
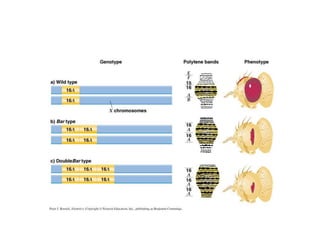
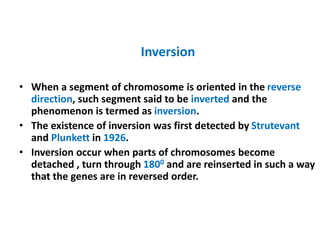
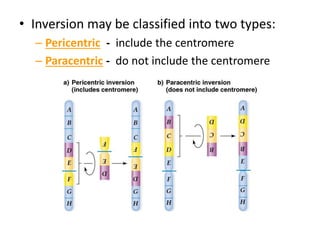
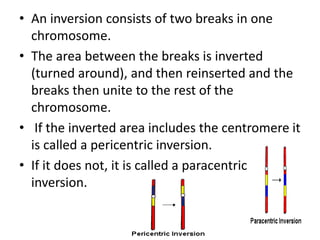
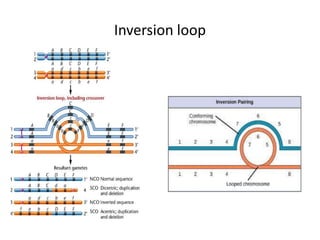
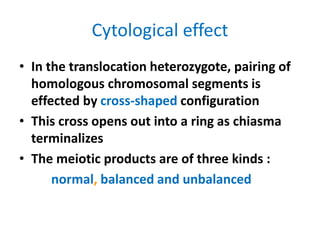
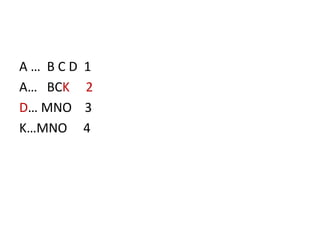
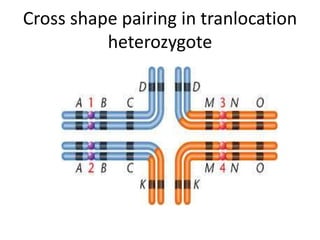
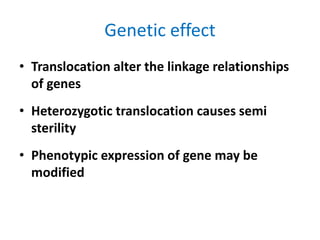
Chromosomal aberrations are variations in chromosome number or structure due to mutations or spontaneous events, categorized mainly into structural and numerical aberrations. Common types of structural abnormalities include deletion, duplication, inversion, and translocation, each affecting the organism's genetic and physiological traits. These aberrations can lead to significant genetic disorders, such as cri-du-chat syndrome and chronic myelocytic leukemia, highlighting their impact on health and development.