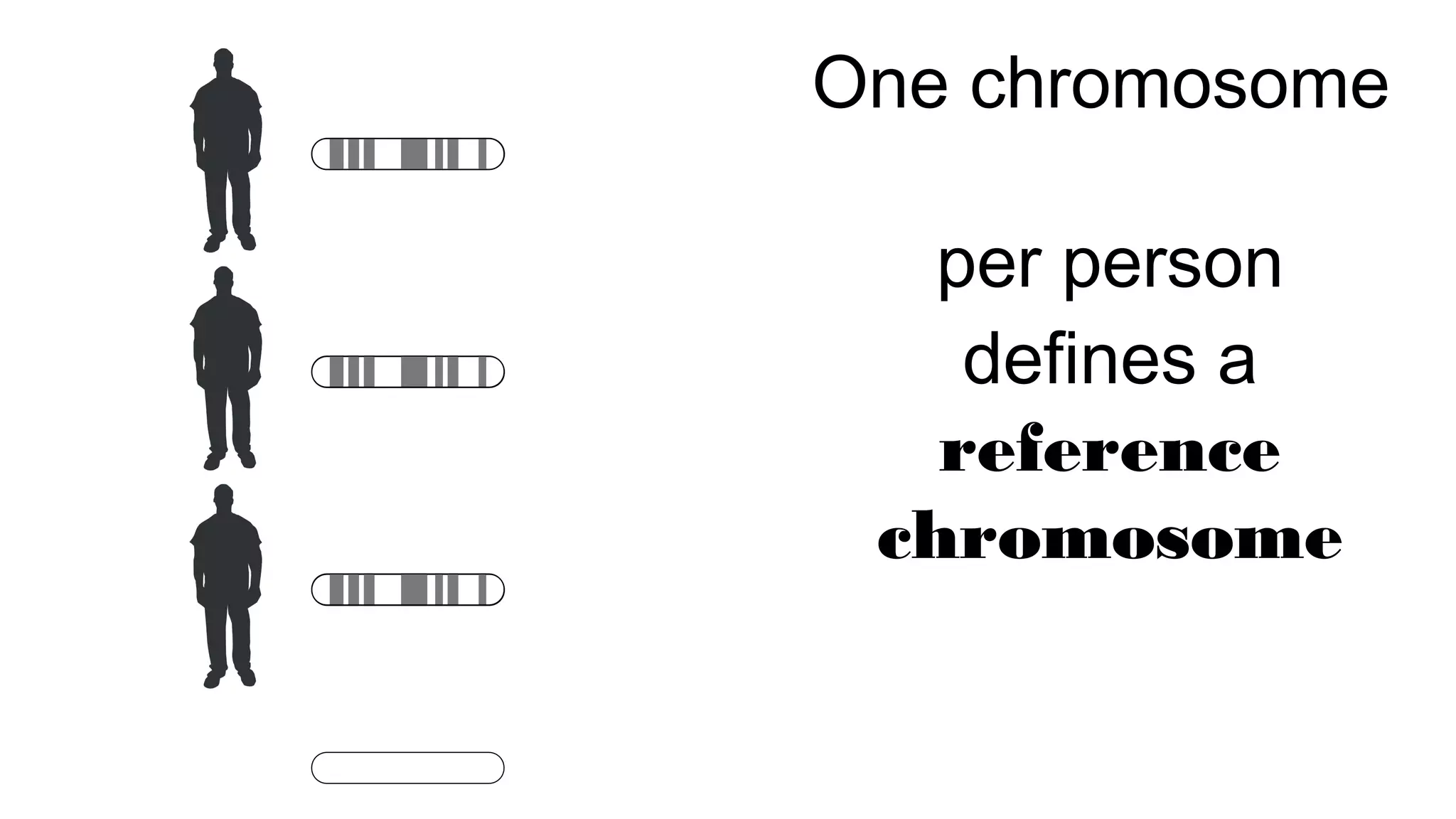
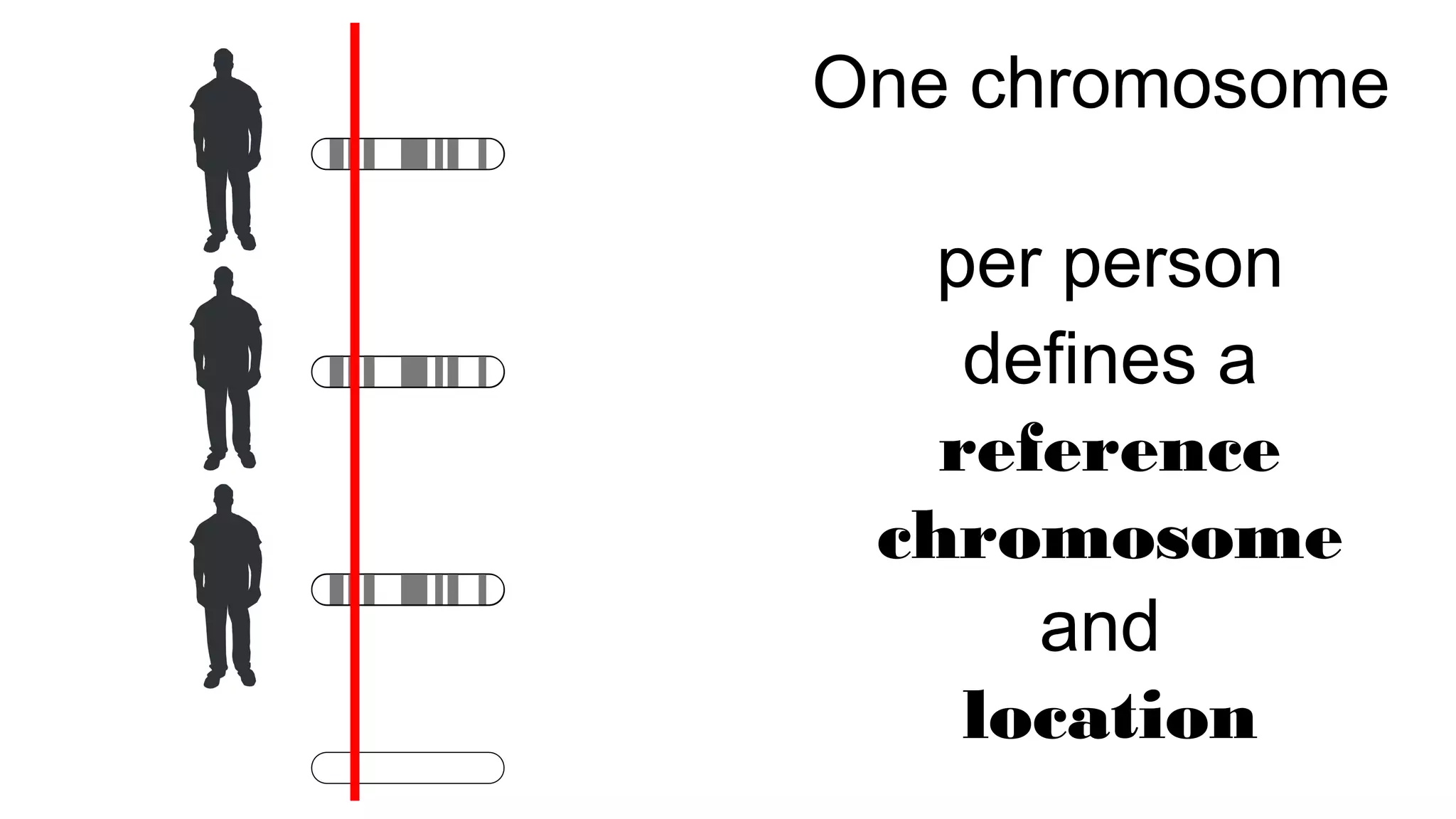
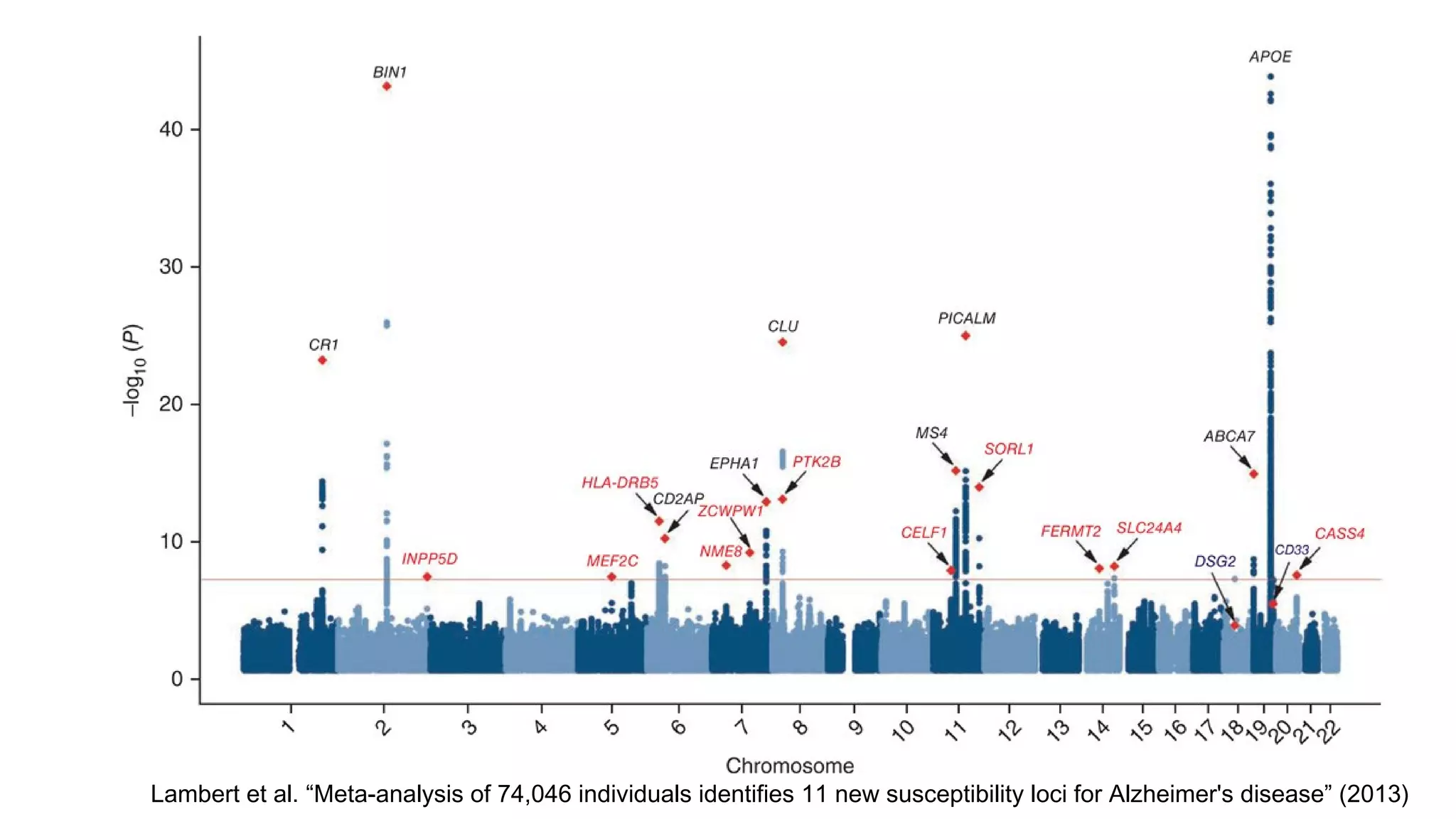
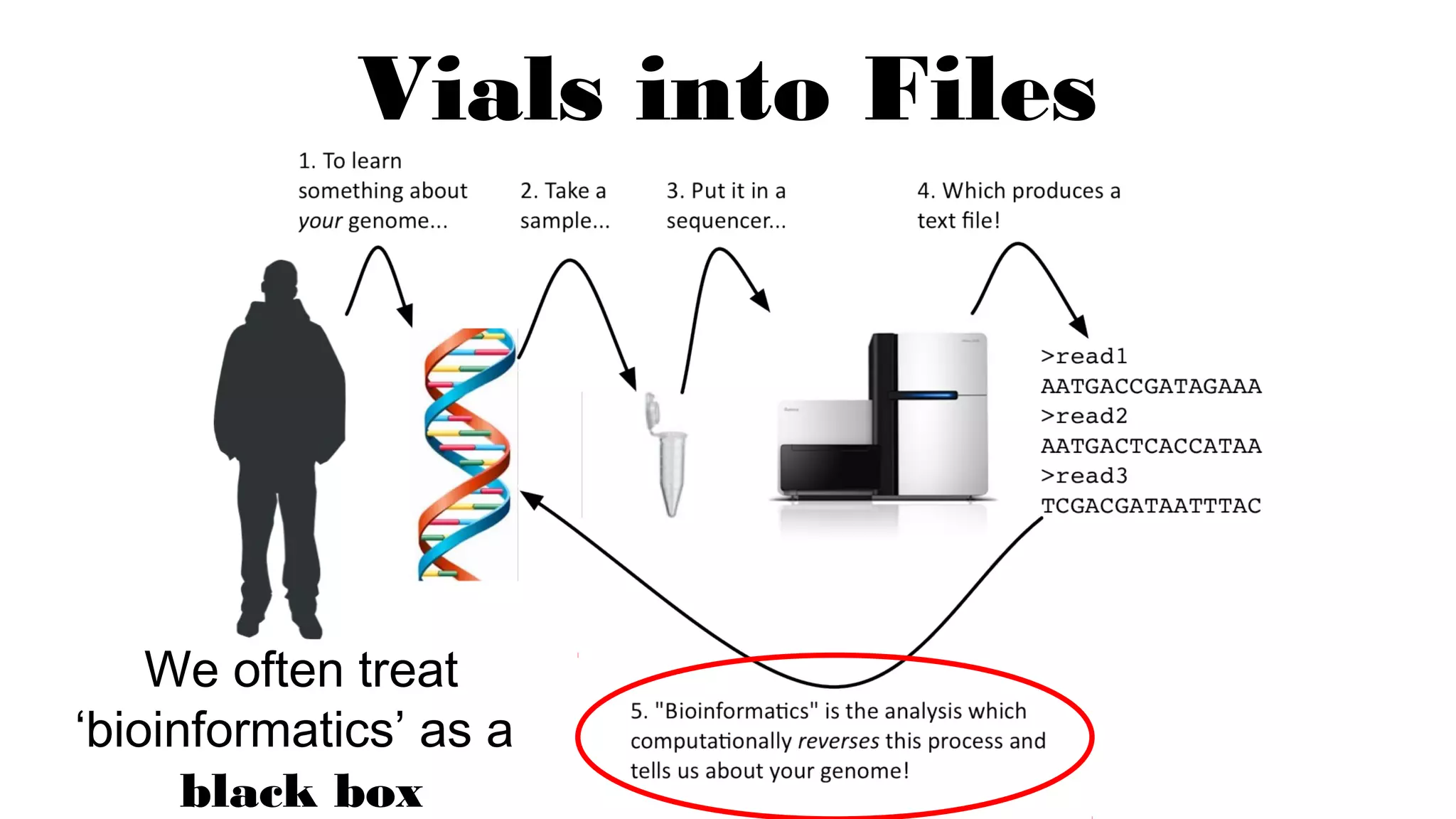
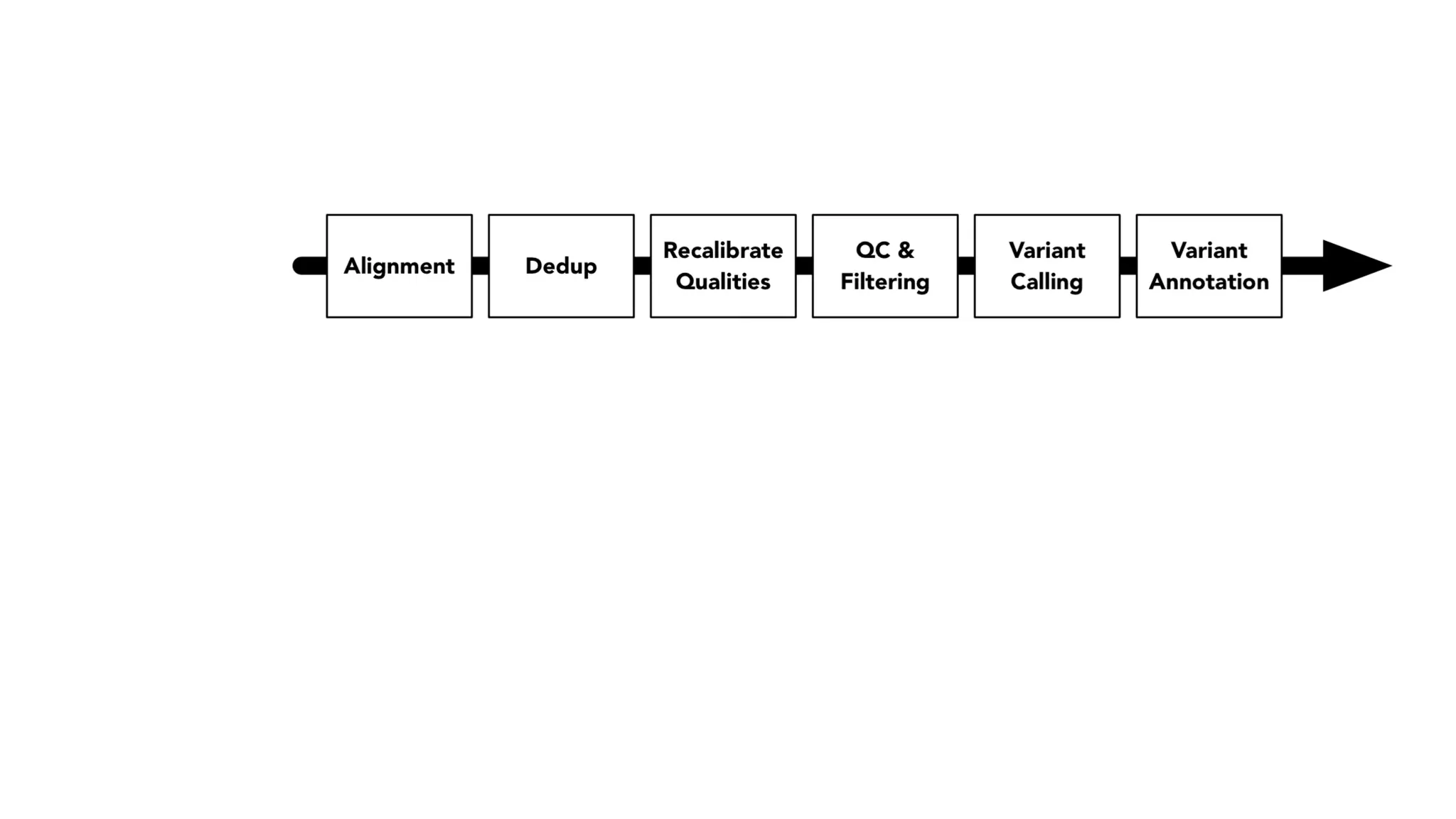
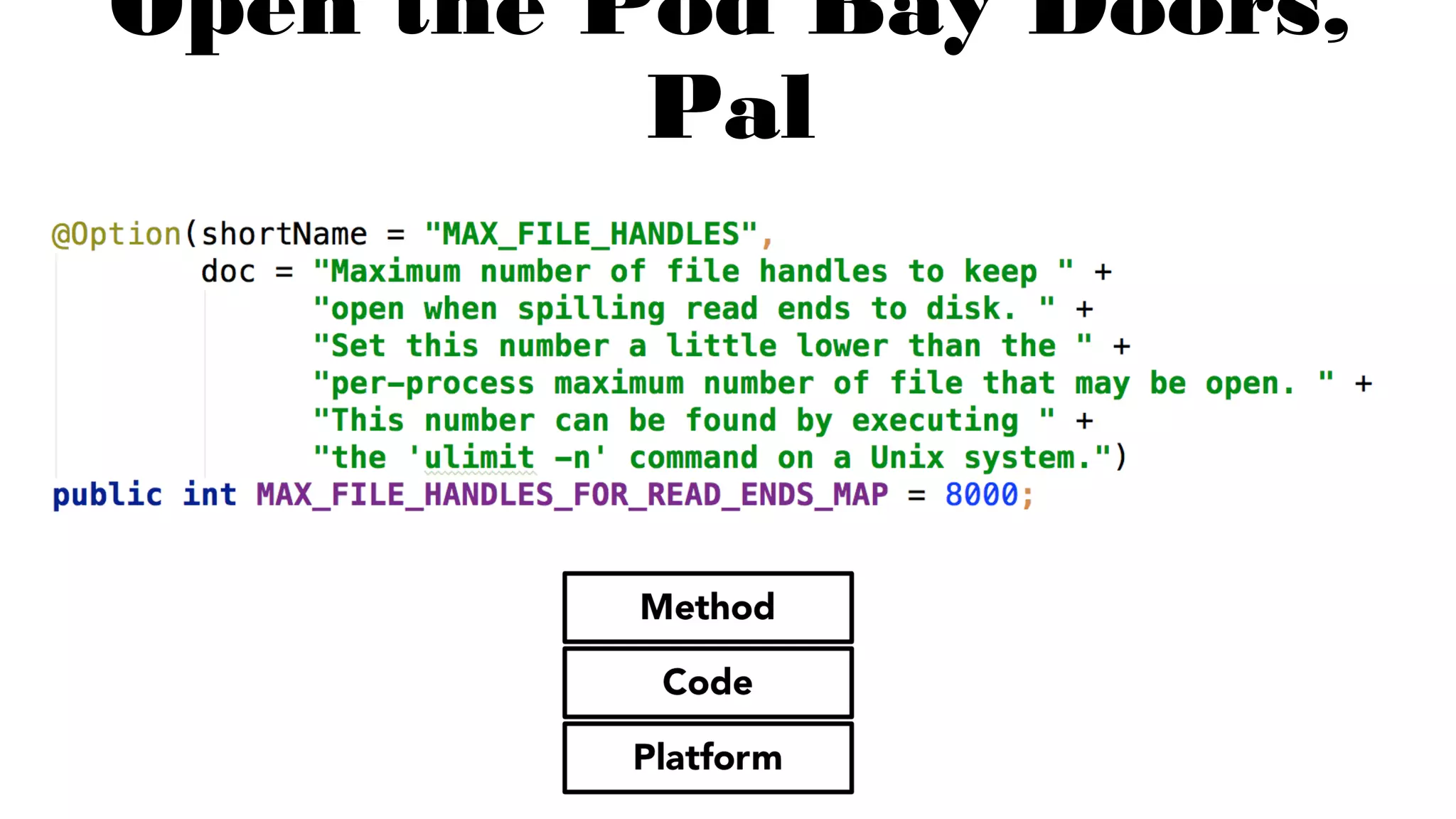
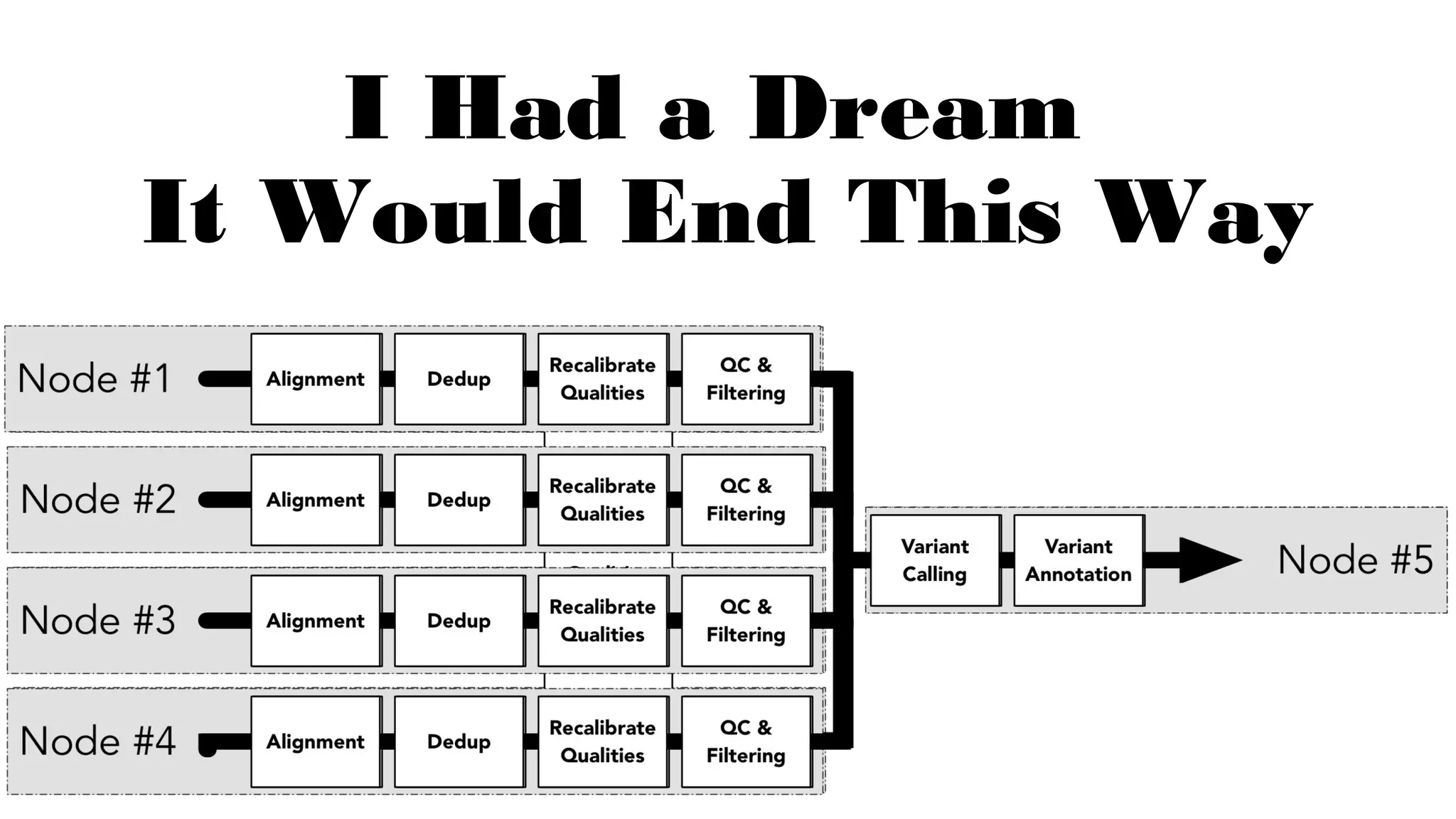
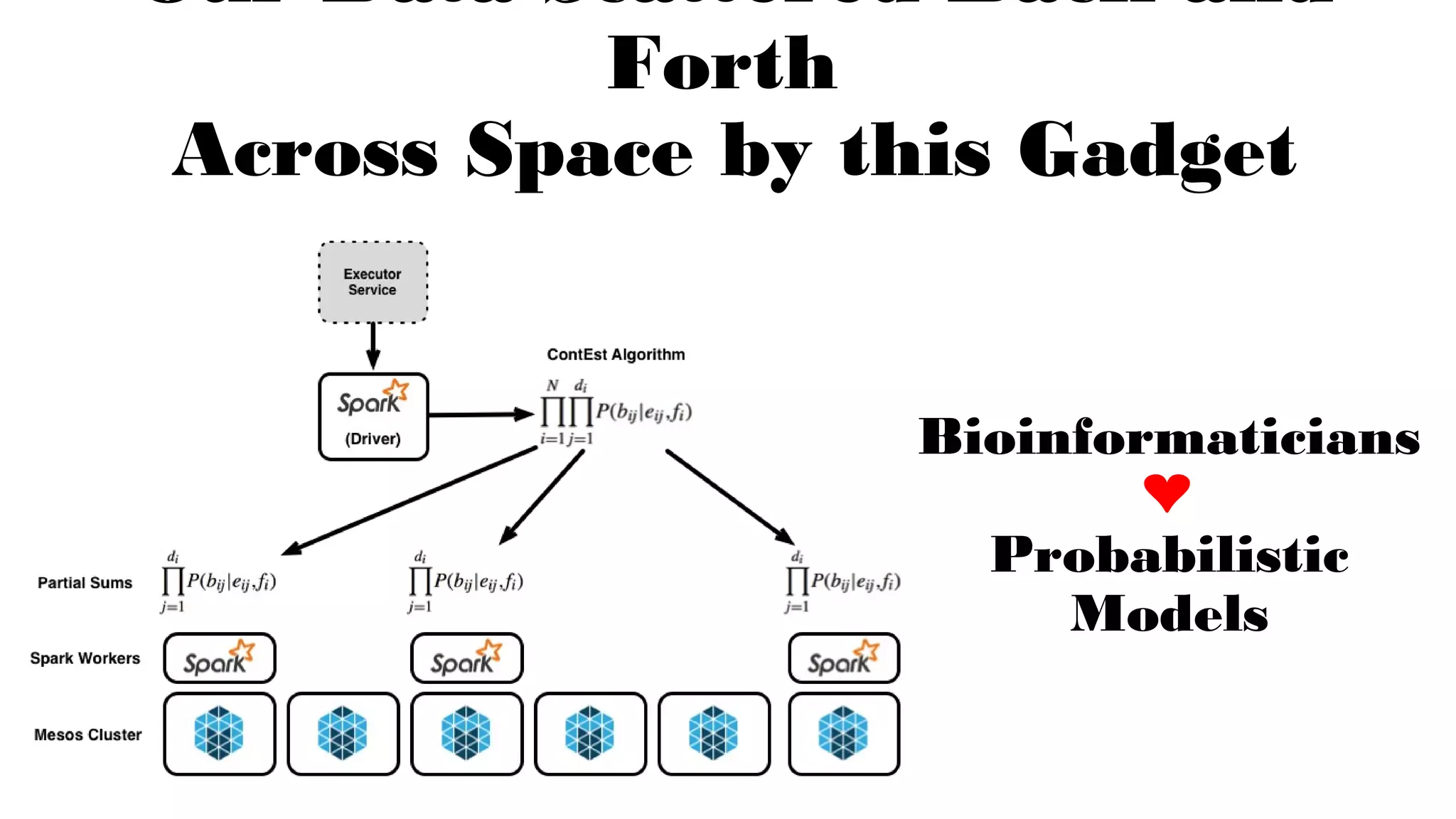
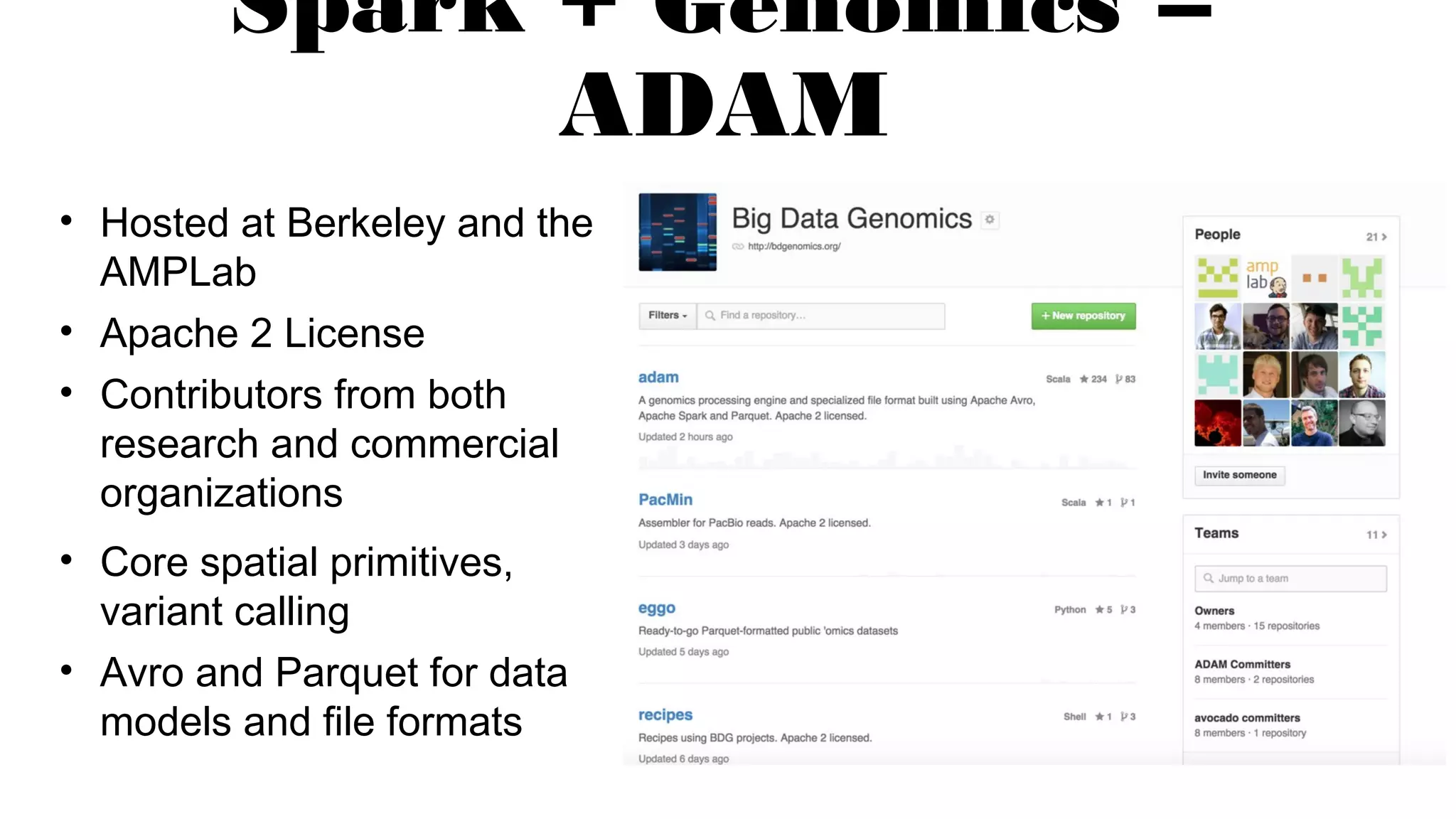
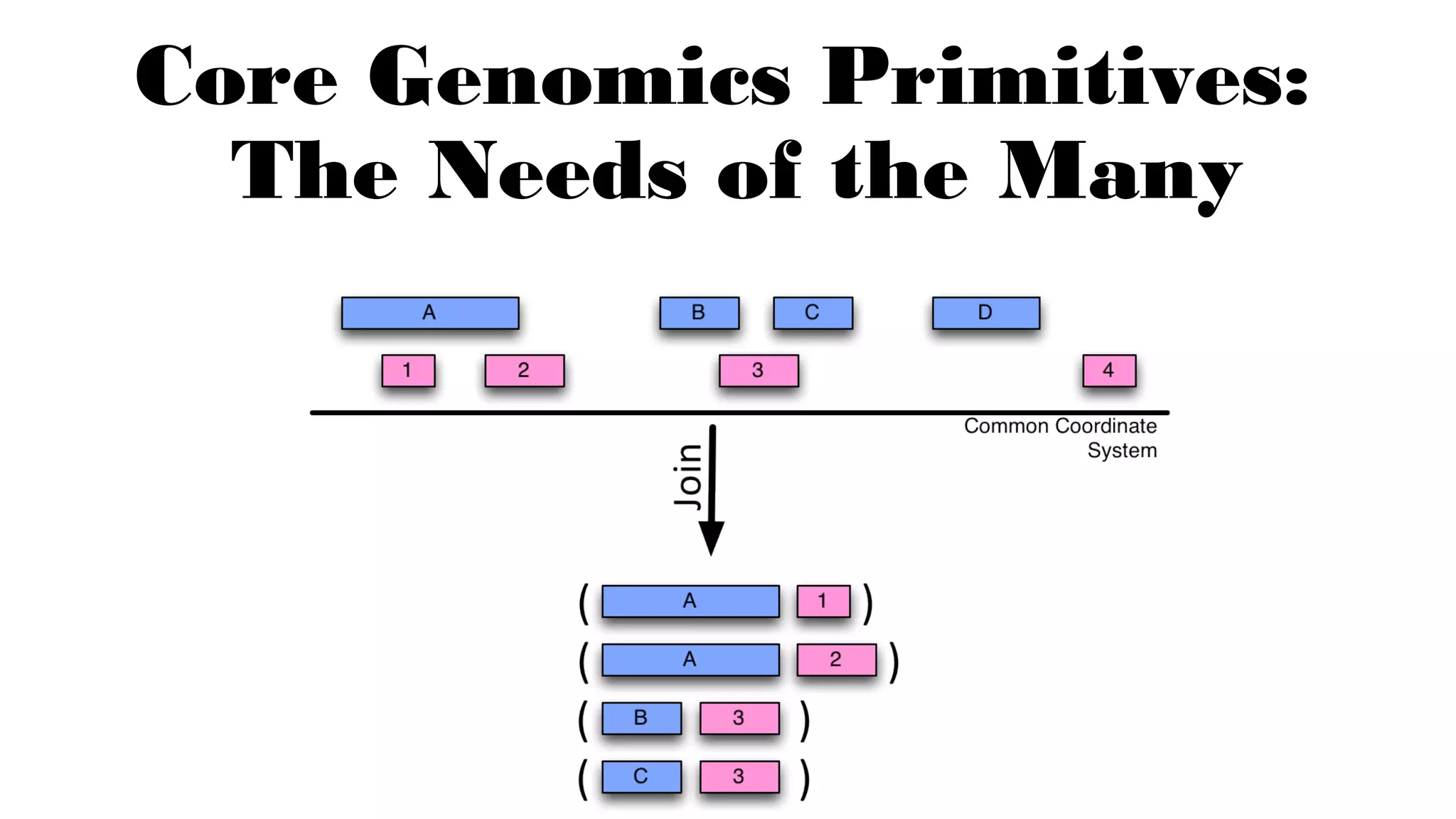
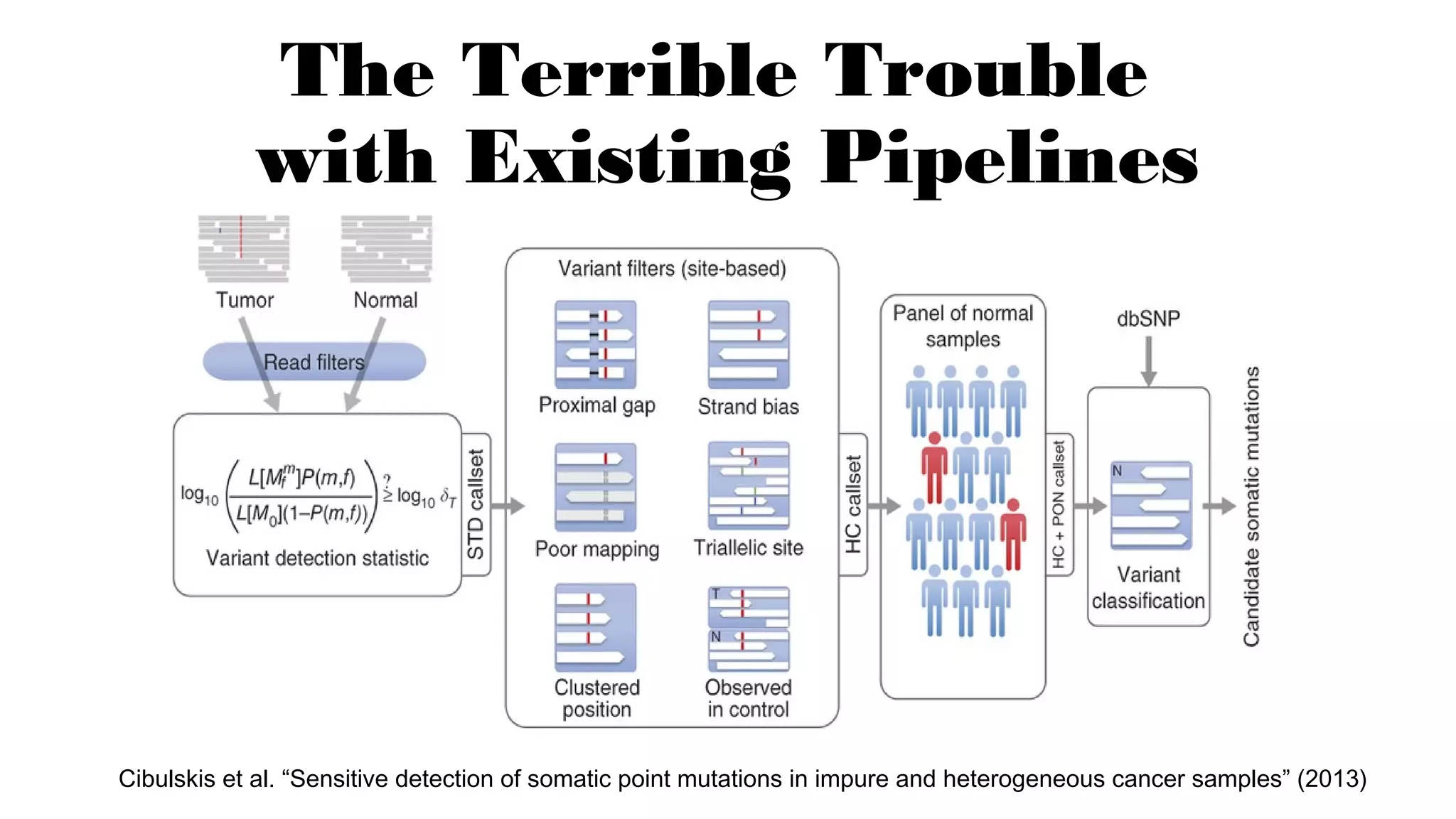
The document discusses advancements in genomics using Spark and ADAM, highlighting the inefficiencies of current bioinformatics pipelines and file formats. It emphasizes the potential improvements of using Parquet for data models and introduces core genomic primitives aided by Spark. The presentation concludes with a critique of common myths surrounding bioinformatics software development.