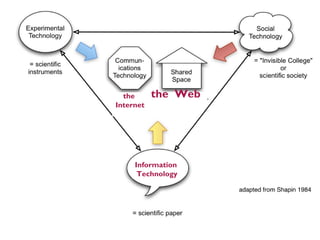
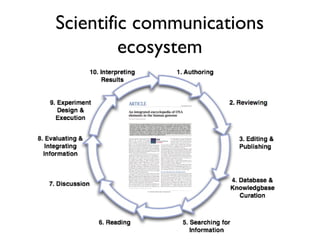
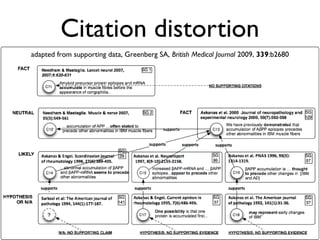
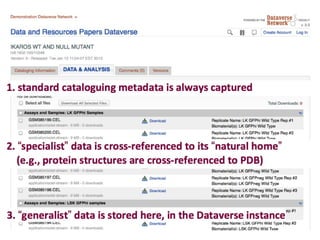
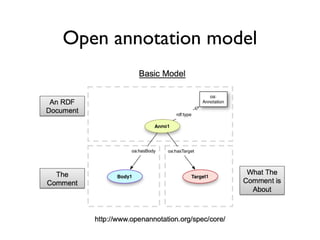
This document discusses challenges with the current scientific publishing system and proposes a vision for next generation scientific publishing (NGSP). Some key problems include retractions due to misconduct, lack of reproducibility, and non-reusable data and methods. NGSP would feature transparent and computable data and methods, open annotation of narratives and objects, and no restrictions on text mining or remixing. It would move information more quickly and allow verification through an open, service-oriented system without walled gardens. Taking NGSP forward will require collaboration across stakeholders in research communications.











![Some problems in the ecosystem
• Intractable publication volumes [1]
• Invalid, distorted and copied citations [3,4,5]
• Growing volume of retractions [5,6]
• 2/3 of retractions due to misconduct [7]
• Research non-reproducibility [8]
• Lack of transparency in publication process [9]
• Methods non-re-usability [10]
• Flawed assessment metrics [11-12]](https://image.slidesharecdn.com/ngsp-130621174607-phpapp01/85/Ngsp-20-320.jpg)












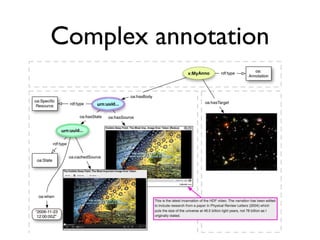



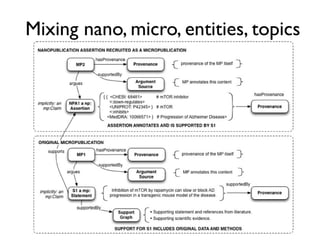
![Digital article summary{
:MP3 rdf:type mp:Micropublication;
mp:name "MP(a3)";
mp:description "Digital summary of Spillman et al. 2010";
pav:authoredBy [ a foaf:Person ; foaf:name "Tim Clark" ];
pav:createdBy [ a foaf:Person ; foaf:name "Tim Clark" ];
pav:createdOn "2013-03-06T09:49:12-05:00"^^xsd:dateTime ;
mp:argues :C3;
mp:supportedBy <info:doi:10.1371/journal.pone.0009979> .
} .
:MP3 = {
:S1 rdf:type mp:Statement;
mp:hasContent "Rapamycin [is] an inhibitor of the mTOR pathway." ;
mp:supportedBy <info:doi/10.1038/nature08221> .
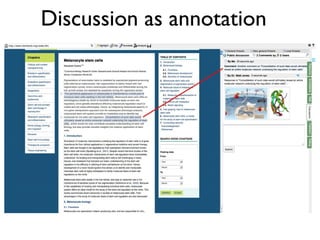
:S2 rdf:type mp:Statement;
mp:hasContent "PDAPP mice accumulate soluble and deposited Aβ and develop AD-like synaptic deficits as well as cognitive
impairment and hippocampal atrophy." ;
mp:supportedBy <info:doi/10.1073/pnas.96.6.3228> .
:S3 rdf:type mp:Statement;
mp:hasContent "Rapamycin-fed transgenic PDAPP mice showed improved learning (Figure 1a) and memory (Figure 1b). We
observed significant deficits in learning and memory in control-fed transgenic PDAPP animals." ;
mp:supportedBy <http://www.jneurosci.org/content/20/11/4050> .
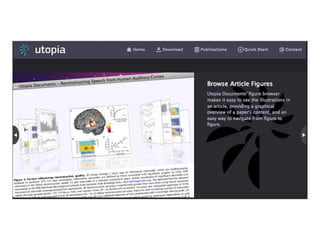
:M1 rdf:type mp:Procedure;
mp:hasName "Rapamycin-supplemented mouse diet protocol" ;
mp:hasContent "We fed a rapamycin-supplemented diet... or control chow to groups of PDAPP mice and littermate non-
transgenic controls for 13 weeks. At the end of treatment (7 mo), learning and memory were tested using the Morris water maze." .
:M2 rdf:type mp:Material;
mp:hasName "PDAPP J20";
mp:hasDescription "Lennart Mucke's PDAPP J20 transgenic mice, as obtained from JAX, stock#006293" ;
mp:describedBy: <http://jaxmice.jax.org/strain/006293.html> .
:D1 rdf:type mp:Data;
pav:retrievedFrom <http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0009979#pone-0009979-g001>;
mp:supportedBy :M1, :M2 .
:C3 rdf:type mp:Claim;
mp:hasContent "Inhibition of mTOR by rapamycin can slow or block AD progression in a transgenic mouse model of the
disease." ;
mp:supportedBy :S1, :S2, :S3, :D1.
} .](https://image.slidesharecdn.com/ngsp-130621174607-phpapp01/85/Ngsp-45-320.jpg)






