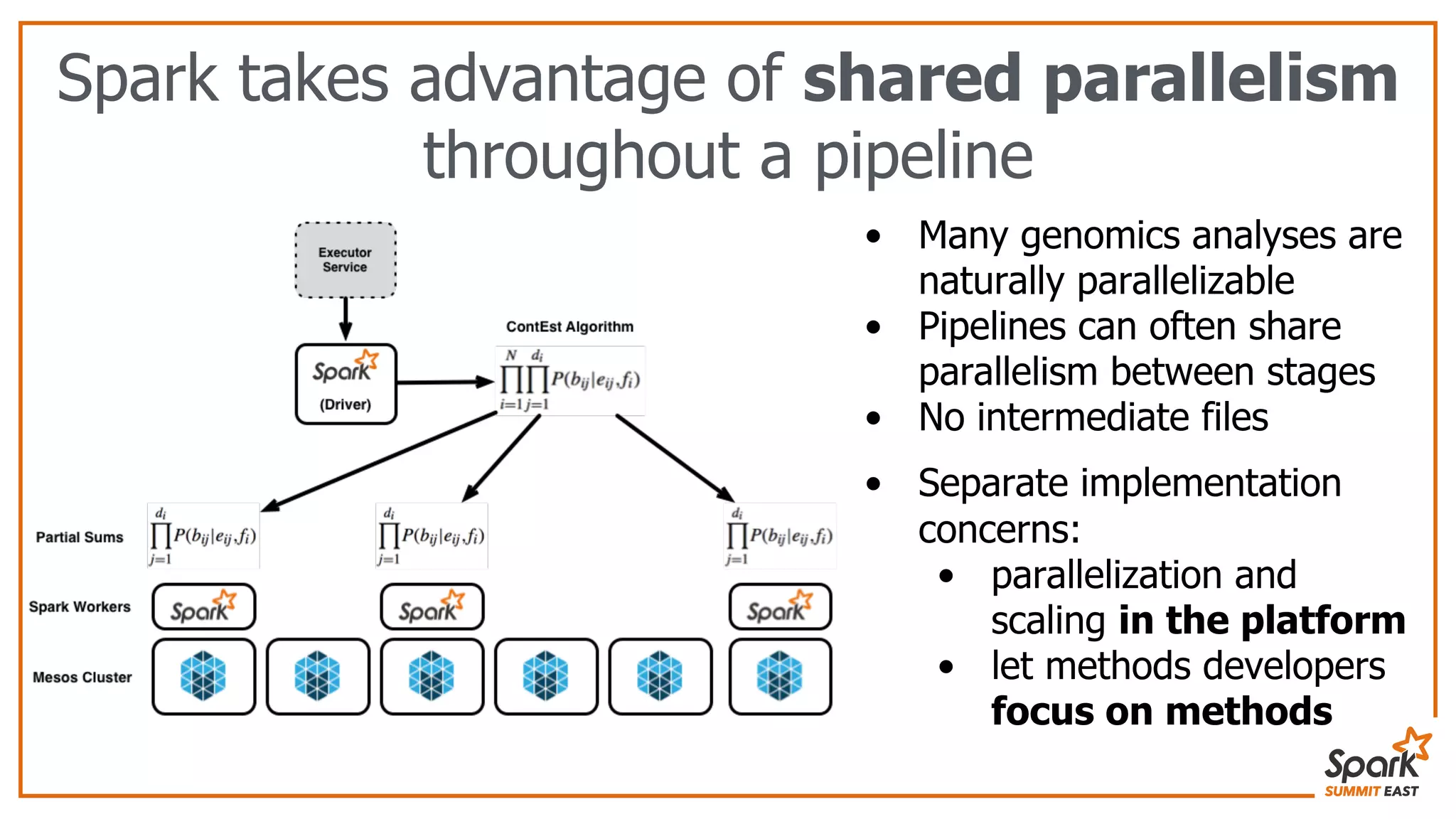
The document discusses the evolution of bioinformatics workflows and the need for scalable, distributed solutions in genomics analysis. It introduces ADAM, a tool built on Apache Spark, which facilitates genomic data processing by leveraging shared parallelism and efficient file formats like Parquet and Avro. The author emphasizes the importance of separating method development from platform building to adapt to the growing complexities of genomic data management.