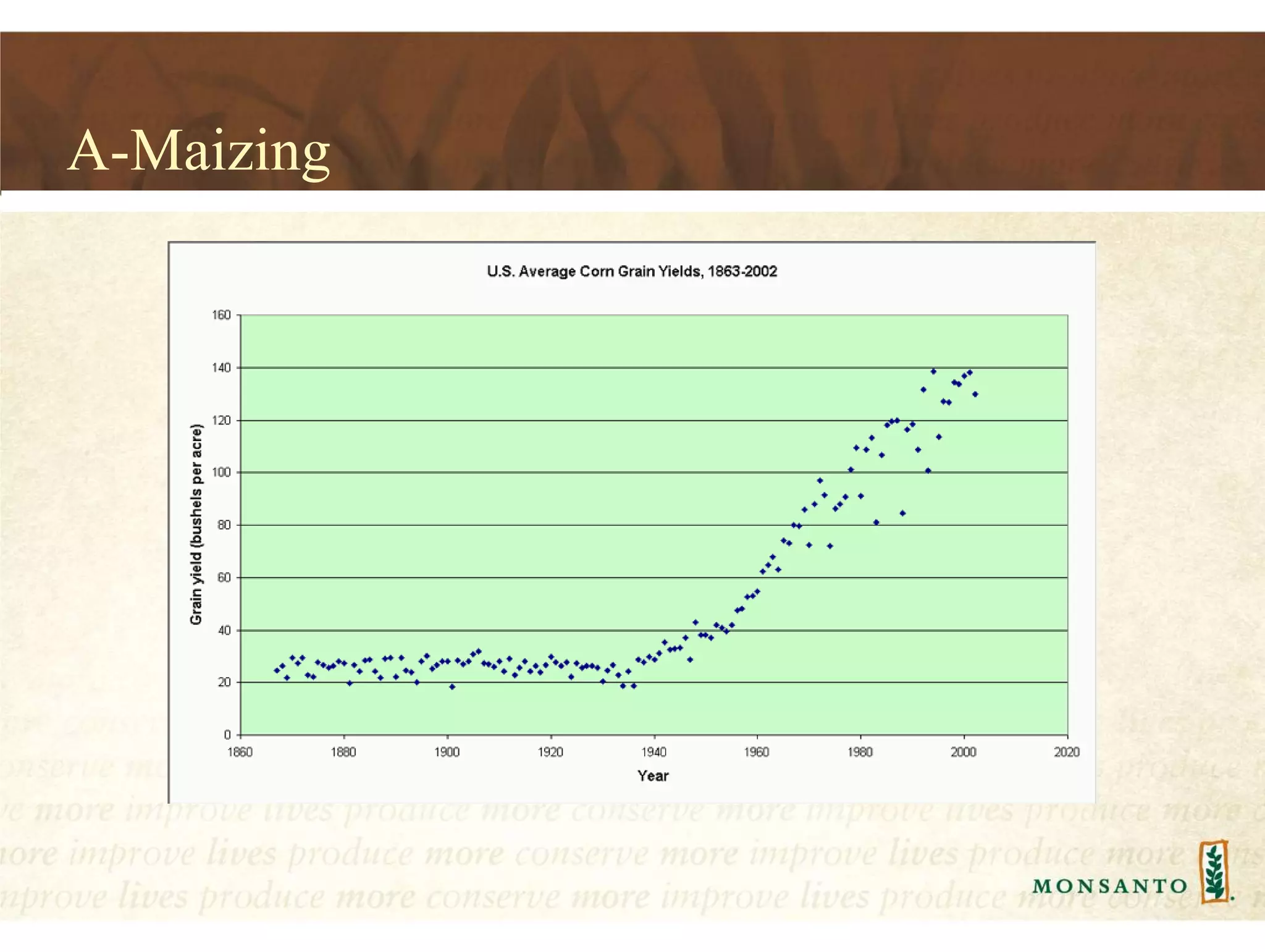
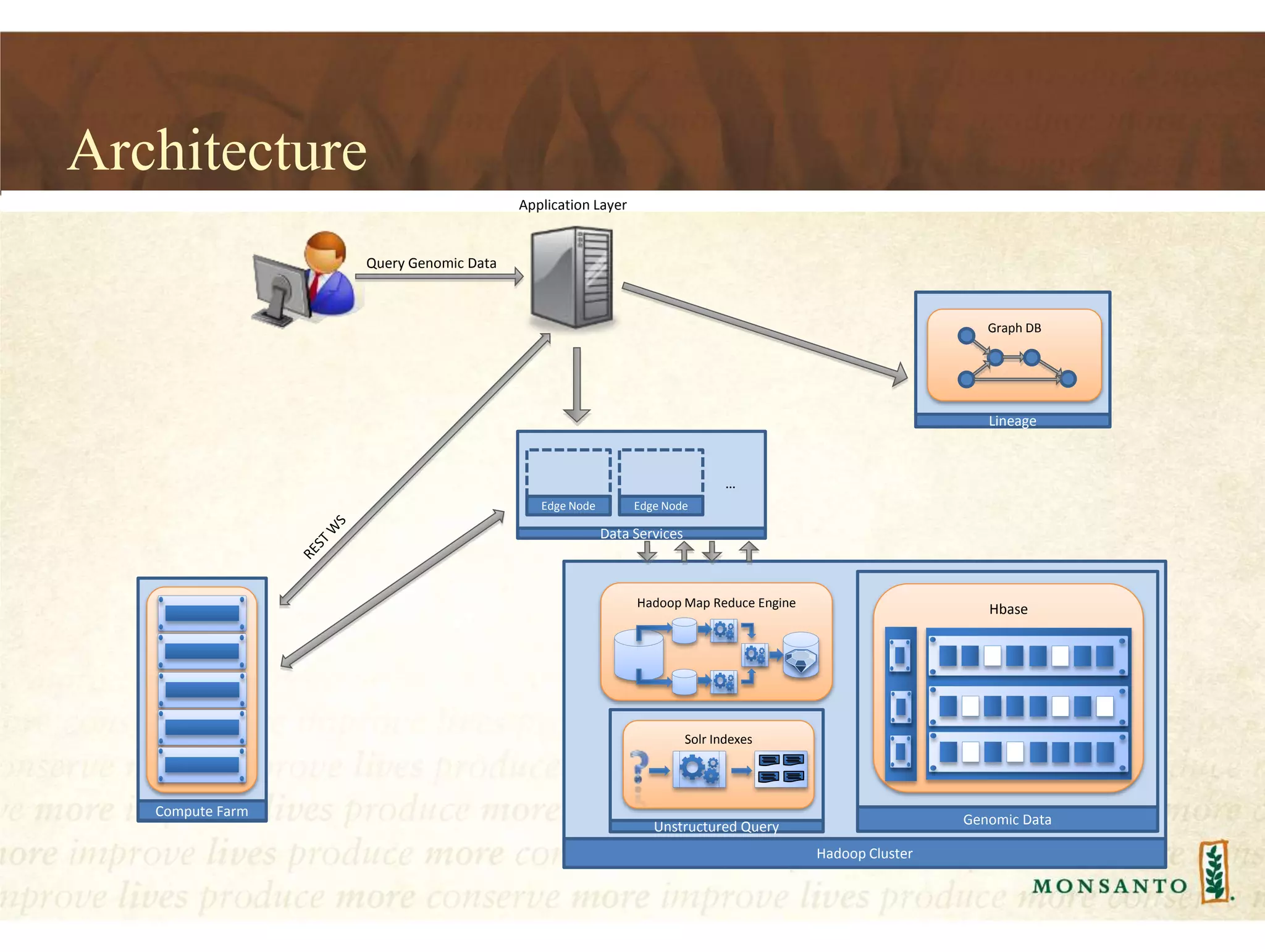
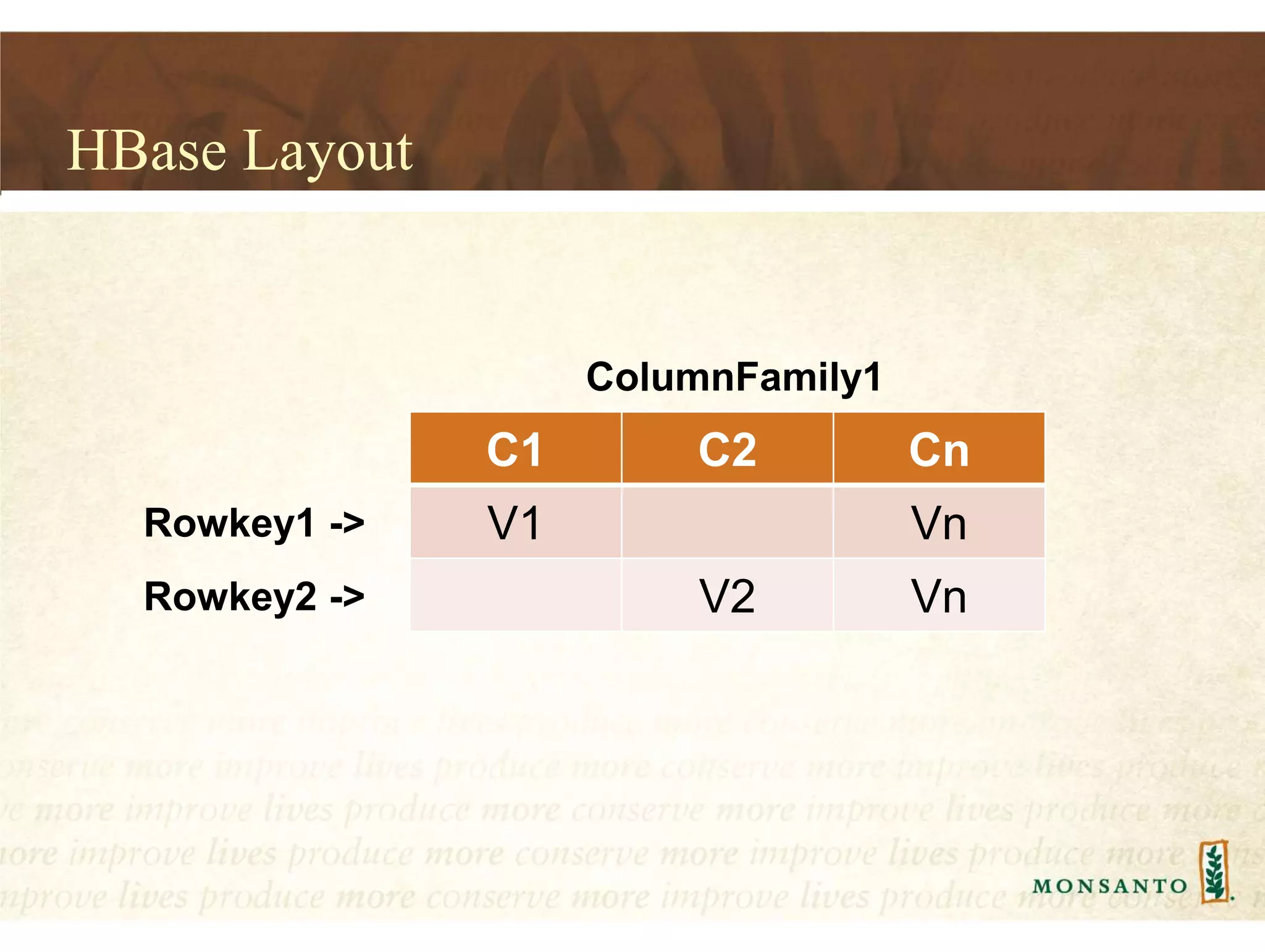
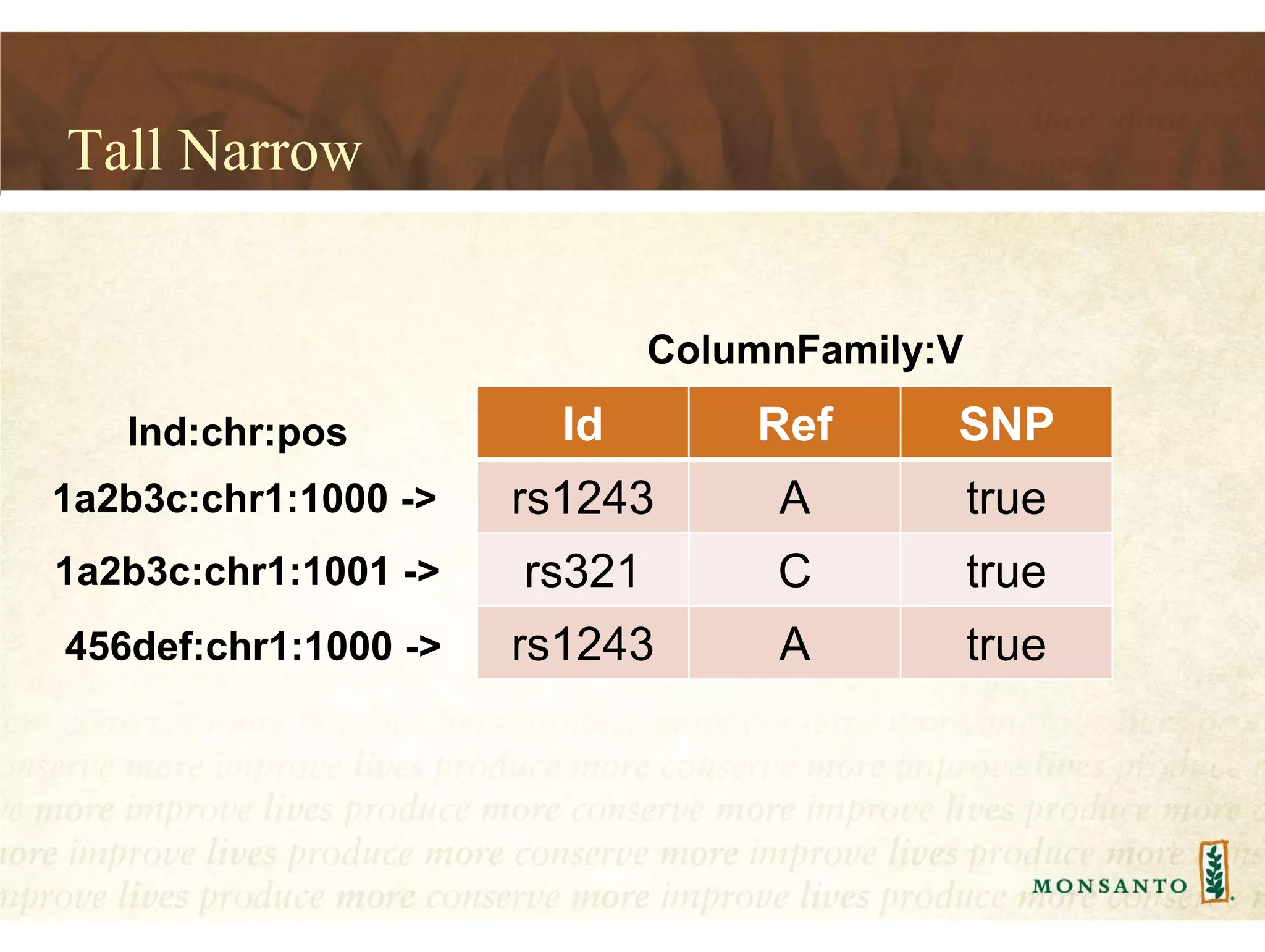
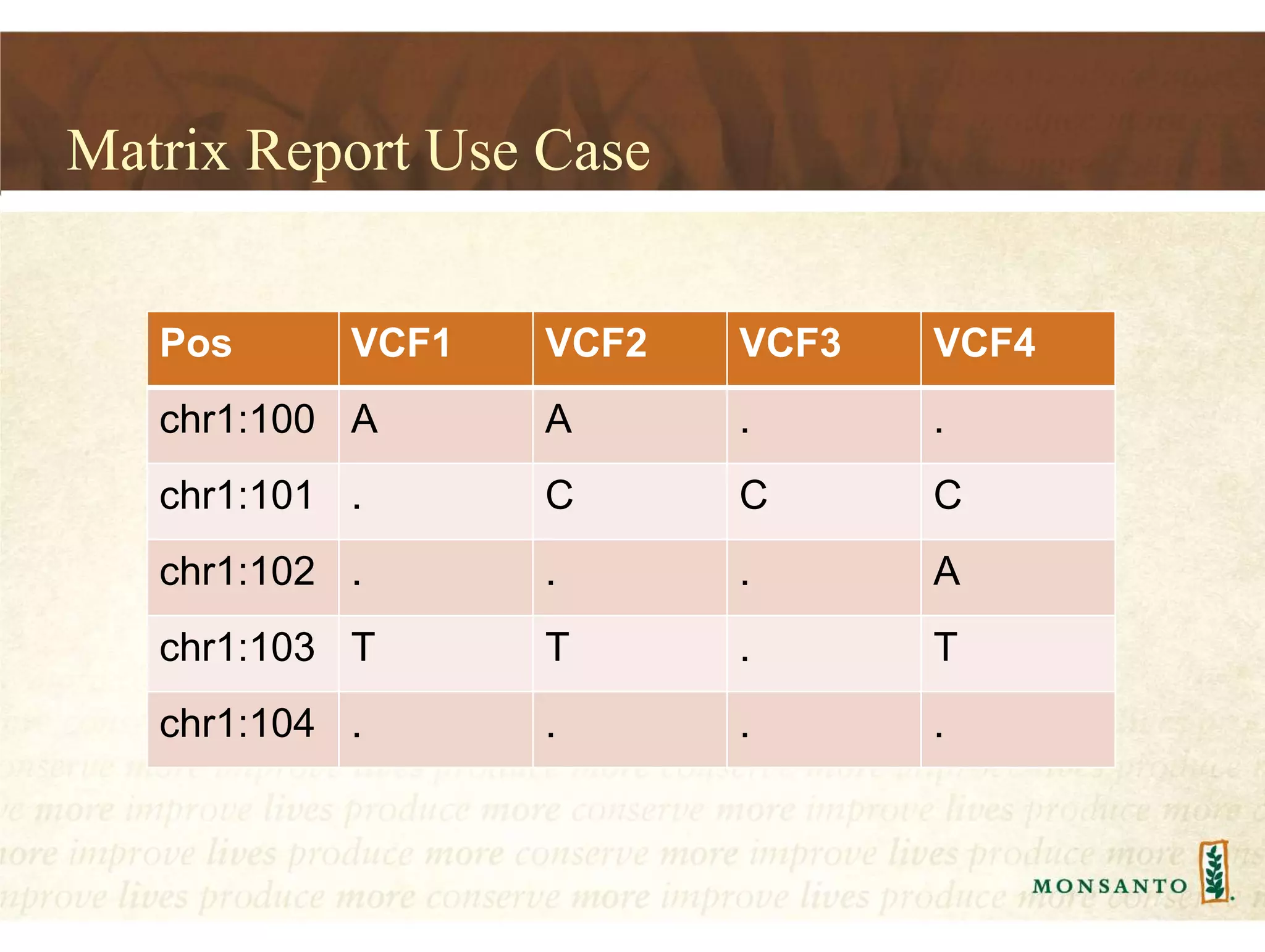
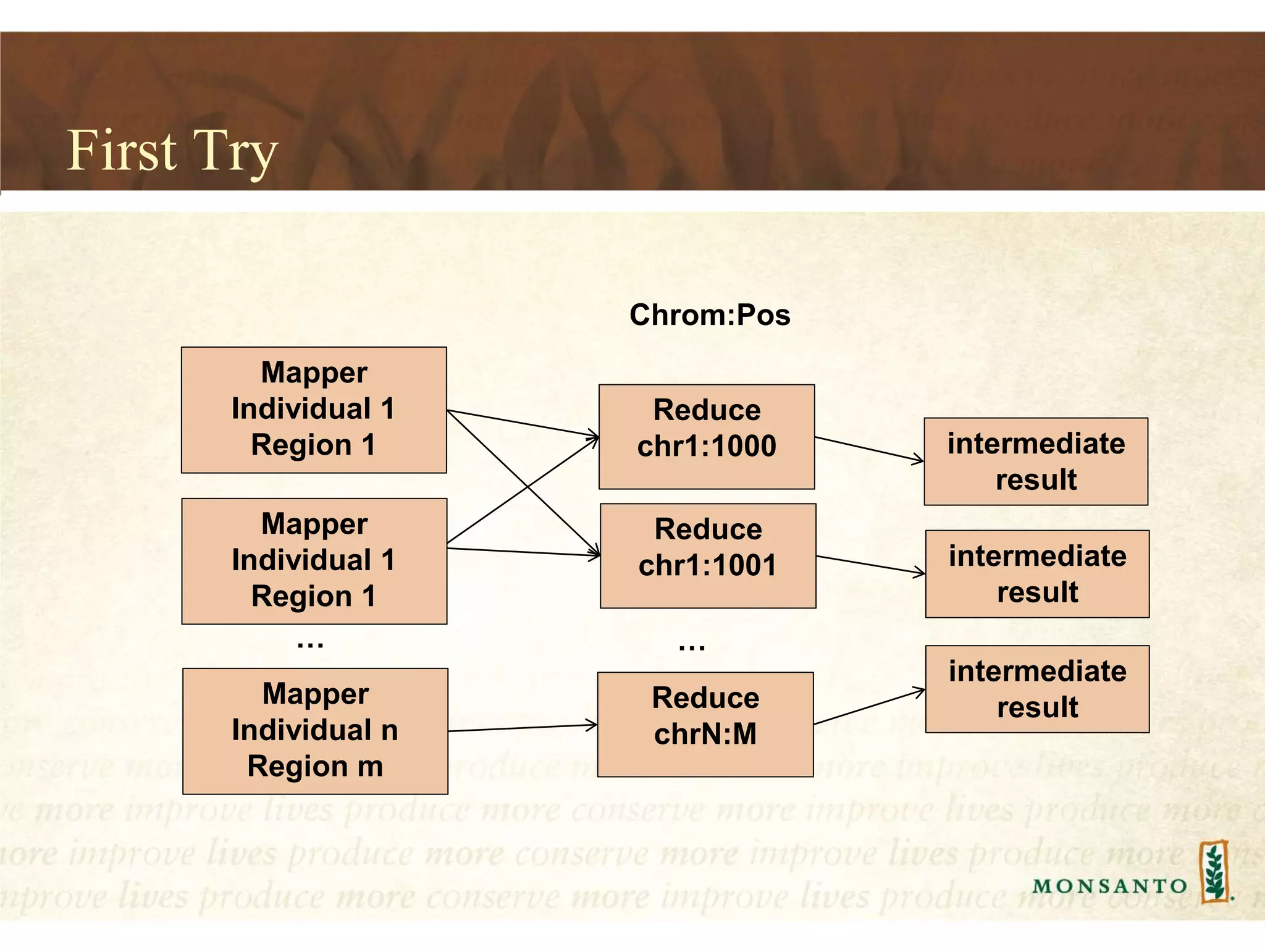
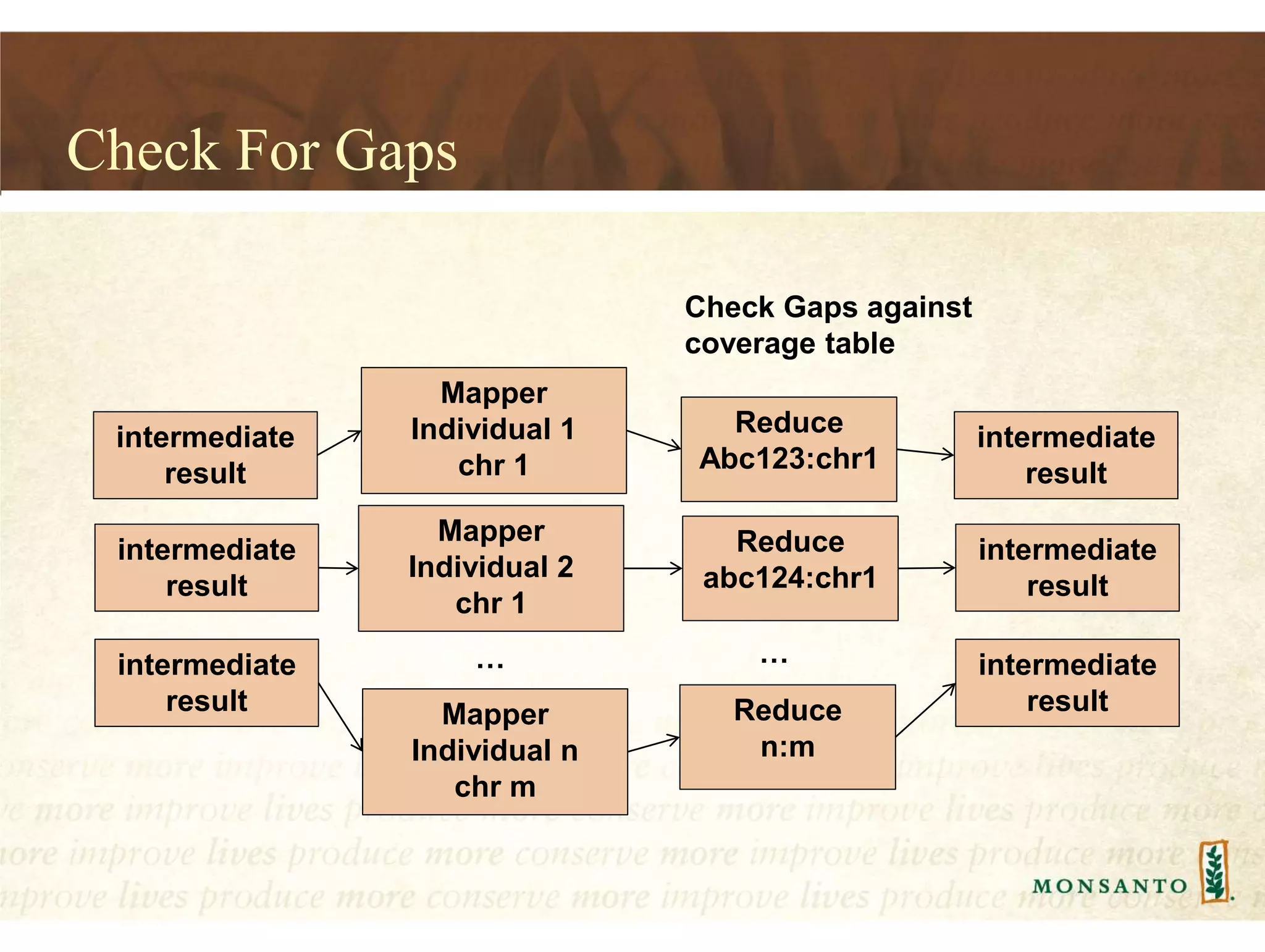
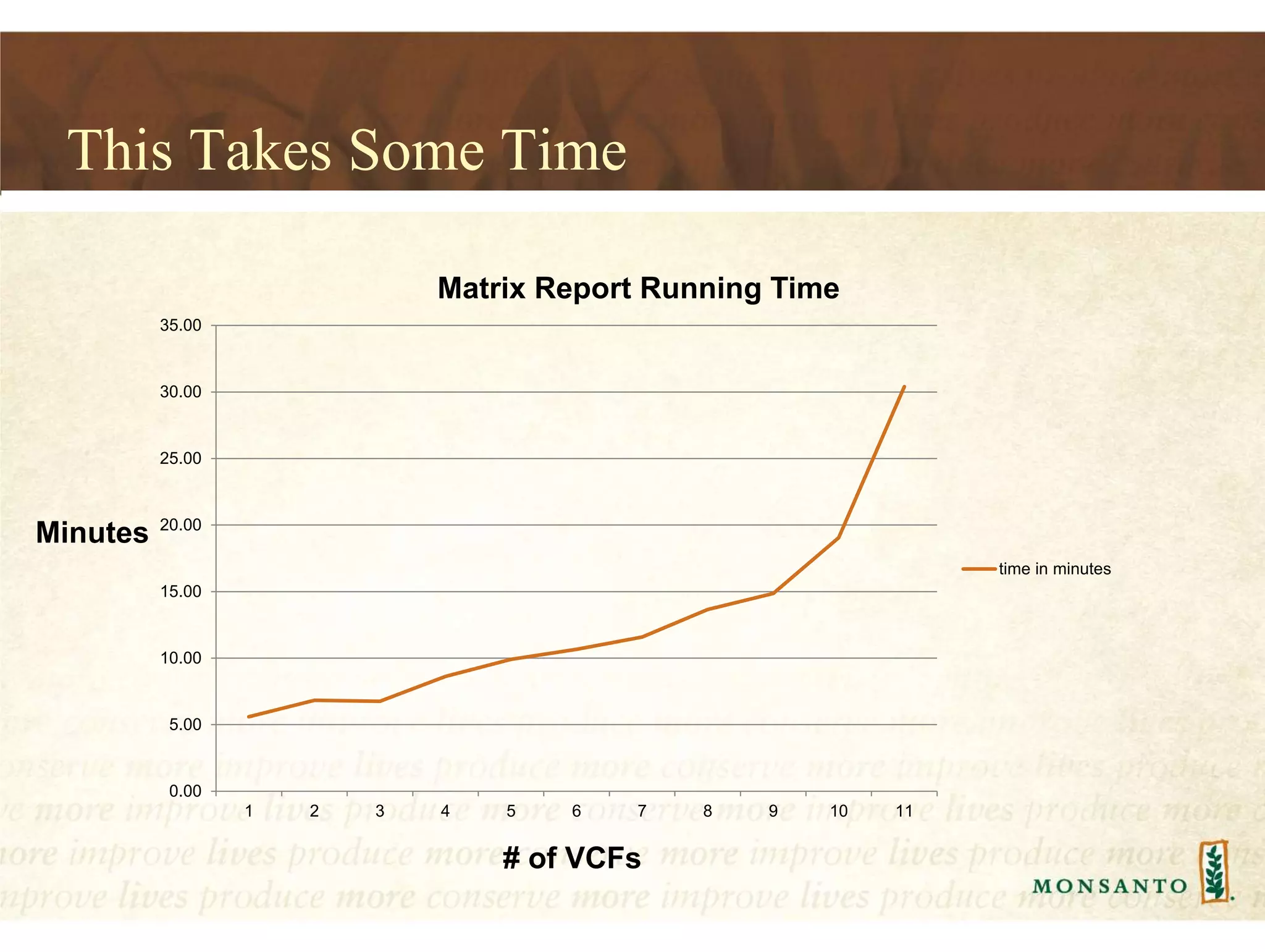
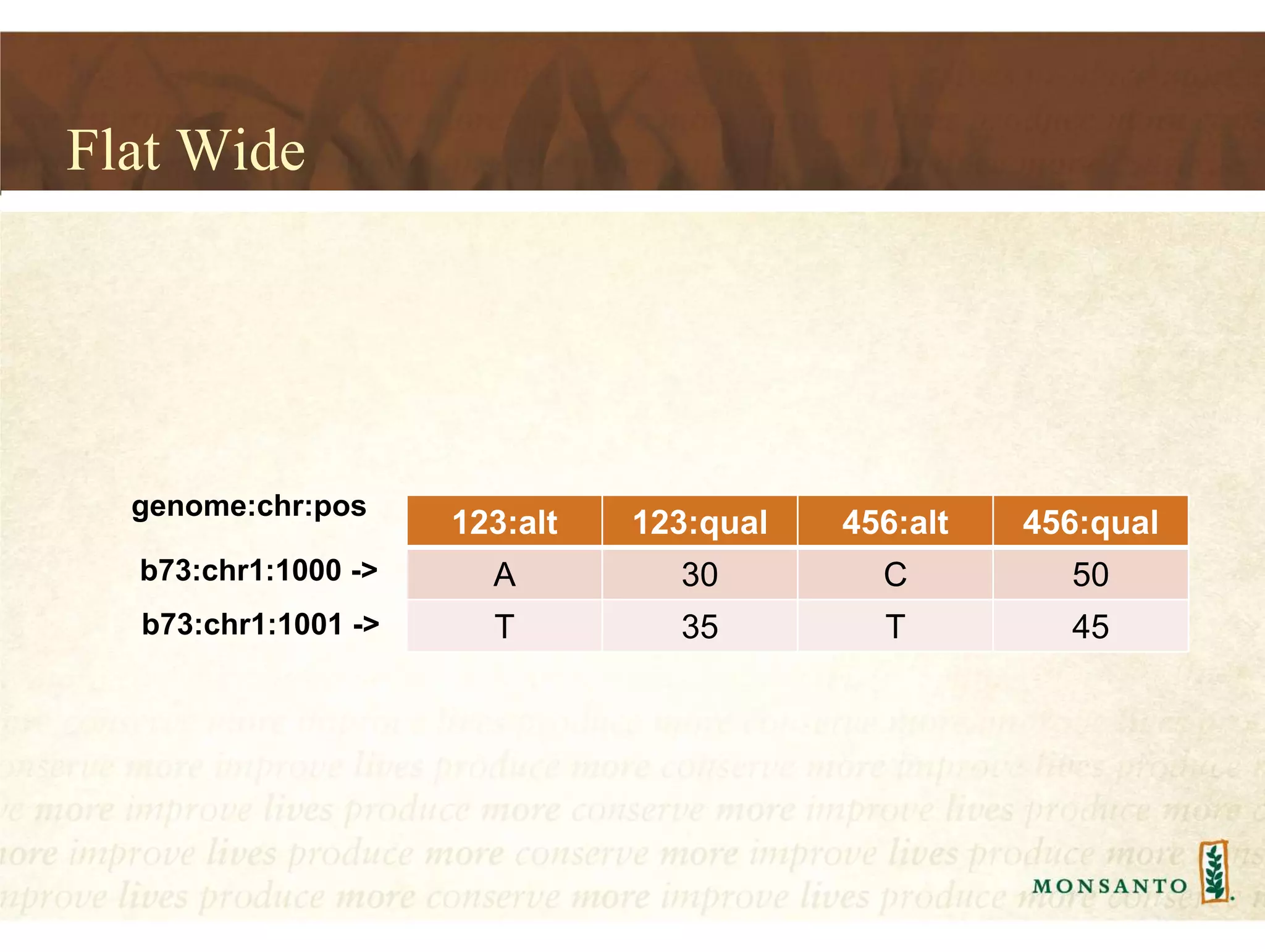
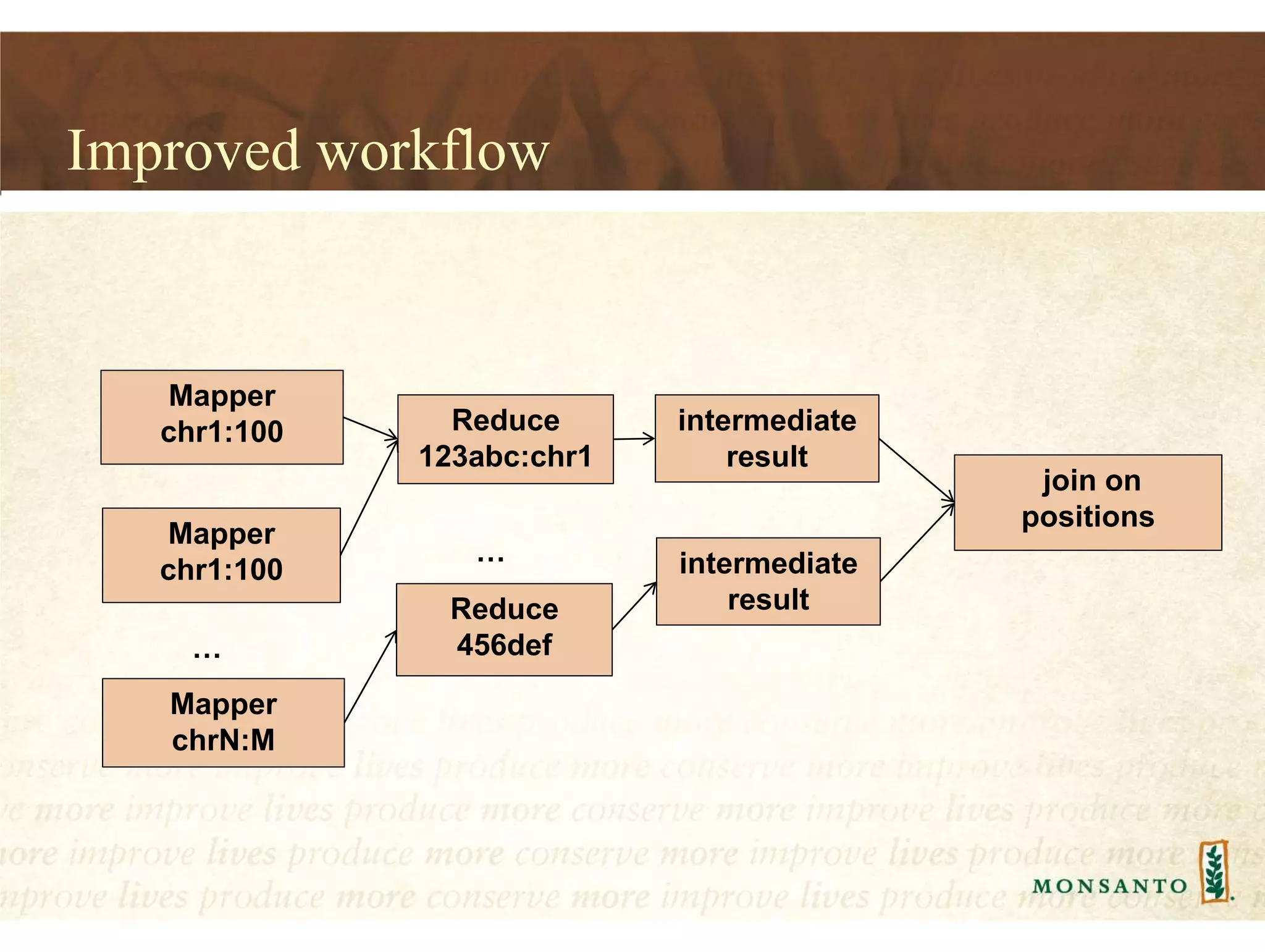
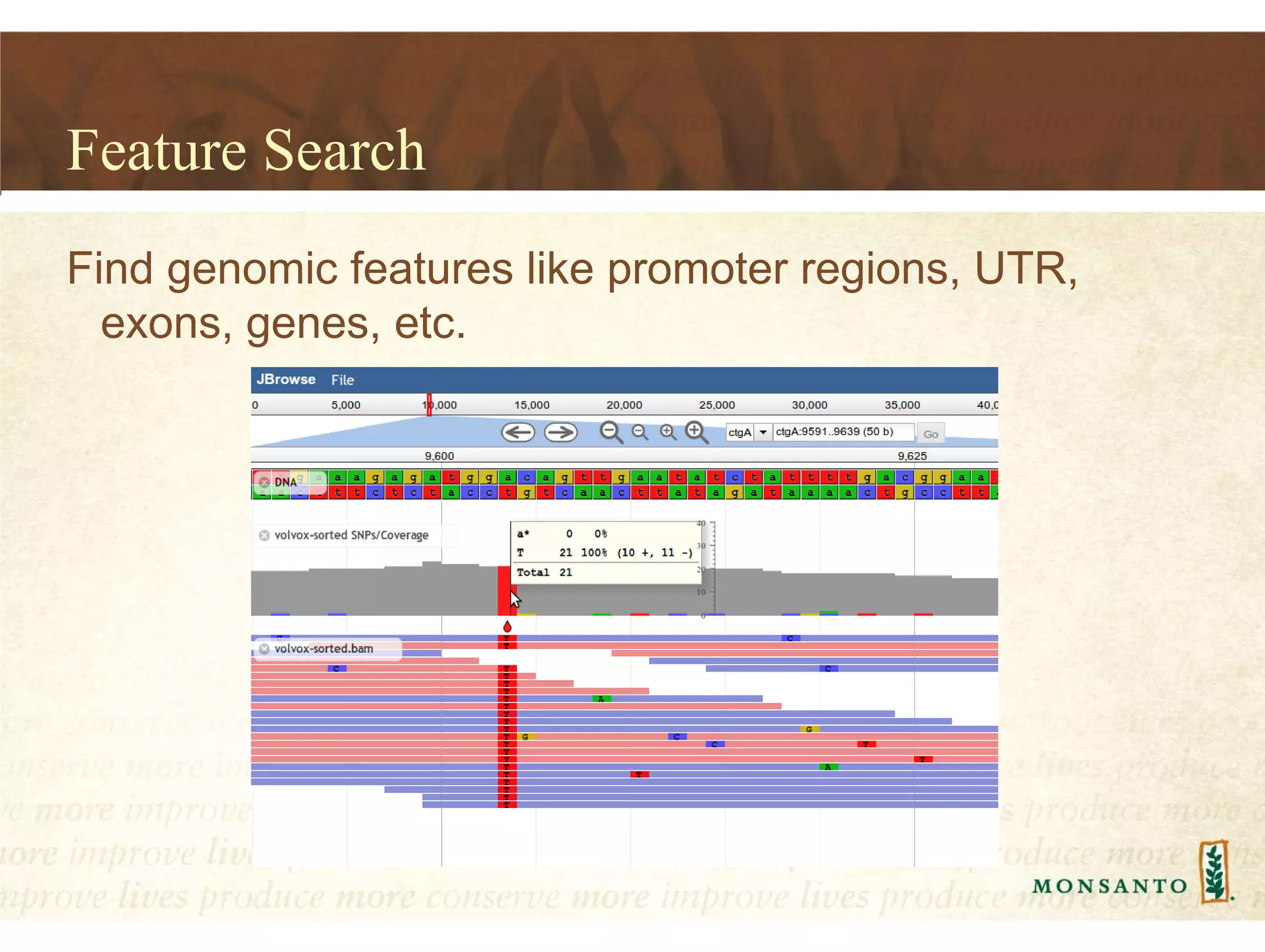
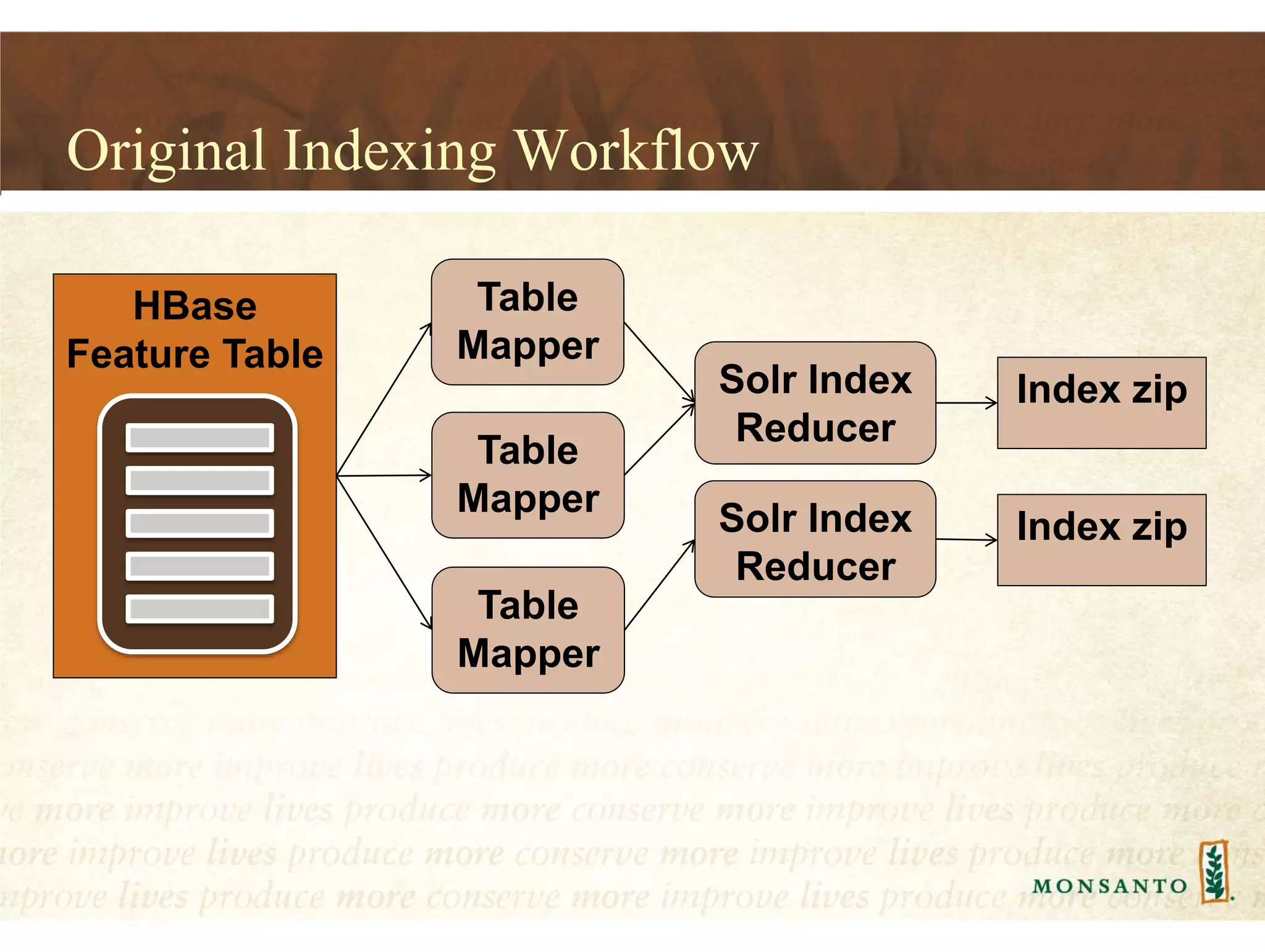
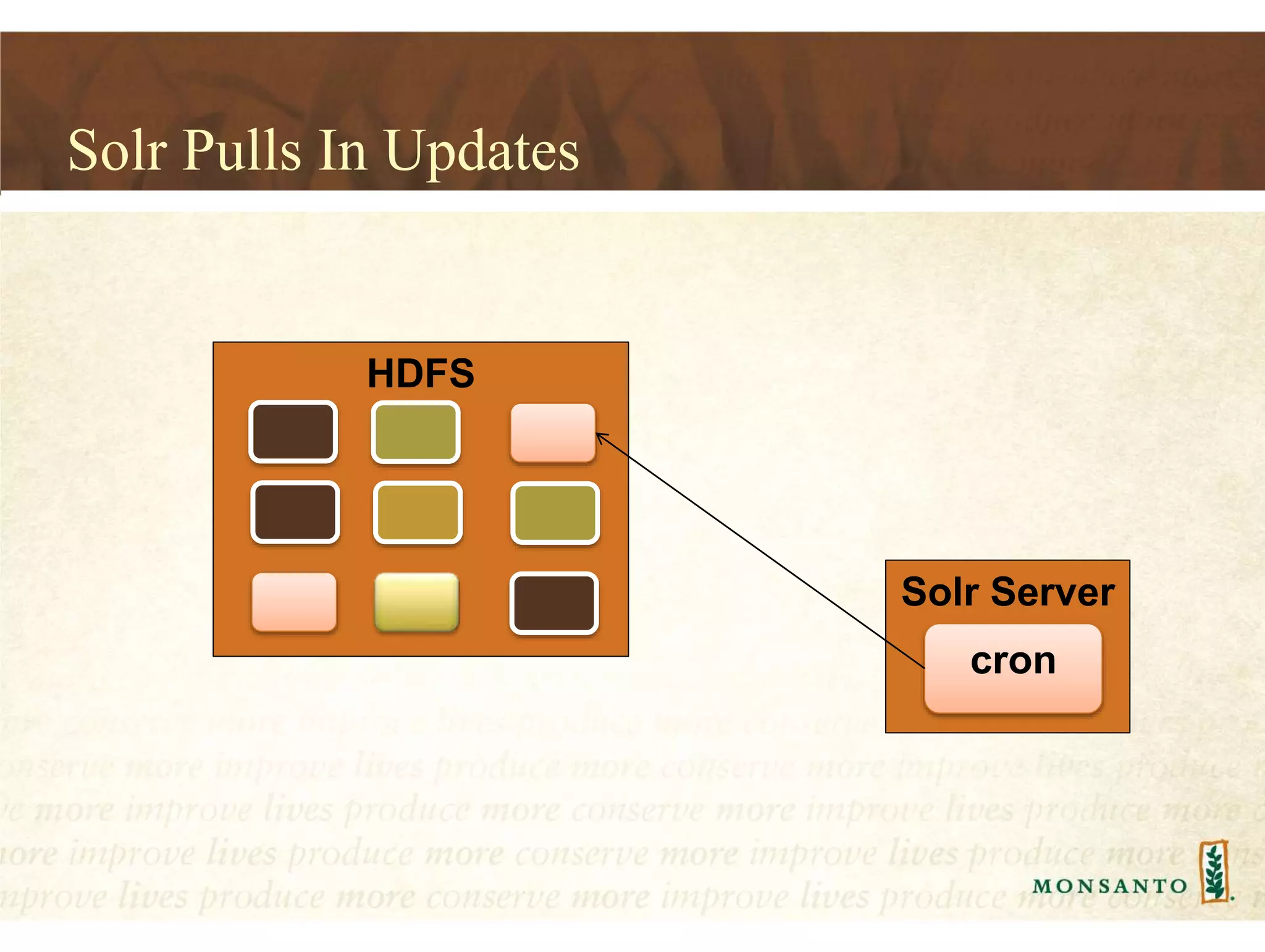
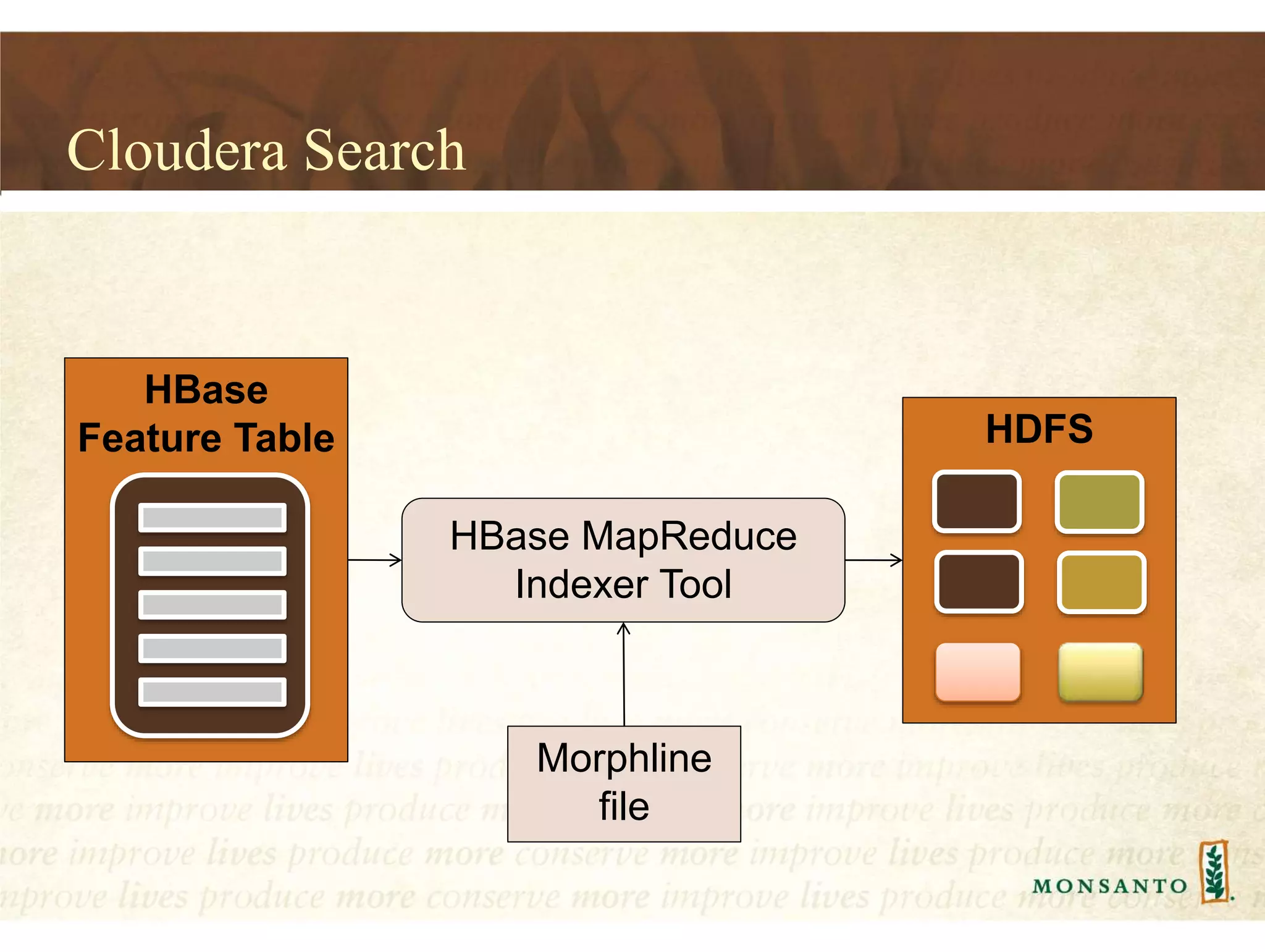
This document summarizes lessons learned from managing large genomic datasets at Monsanto. It discusses how Monsanto uses big data technologies like Hadoop, HBase, and Solr to store and query genomic data at scale. Key lessons include using HBase more like a hashmap than a relational database, denormalizing HBase schemas, and using distributed search technologies like SolrCloud rather than rebuilding Solr indexes. The document provides examples of genomic data formats and architectures used to store, index, and retrieve genomic feature data from petabytes of sequence data.