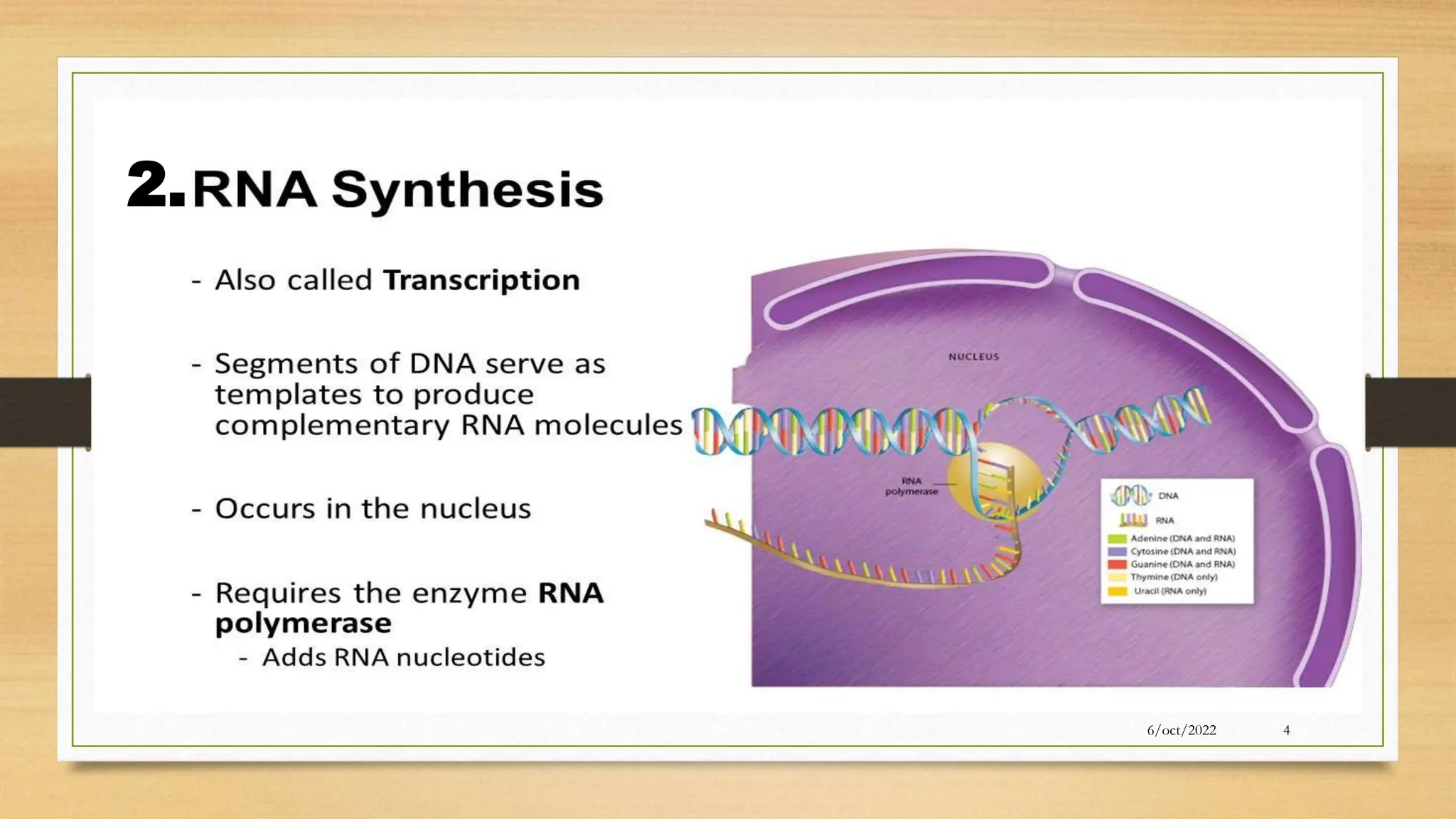
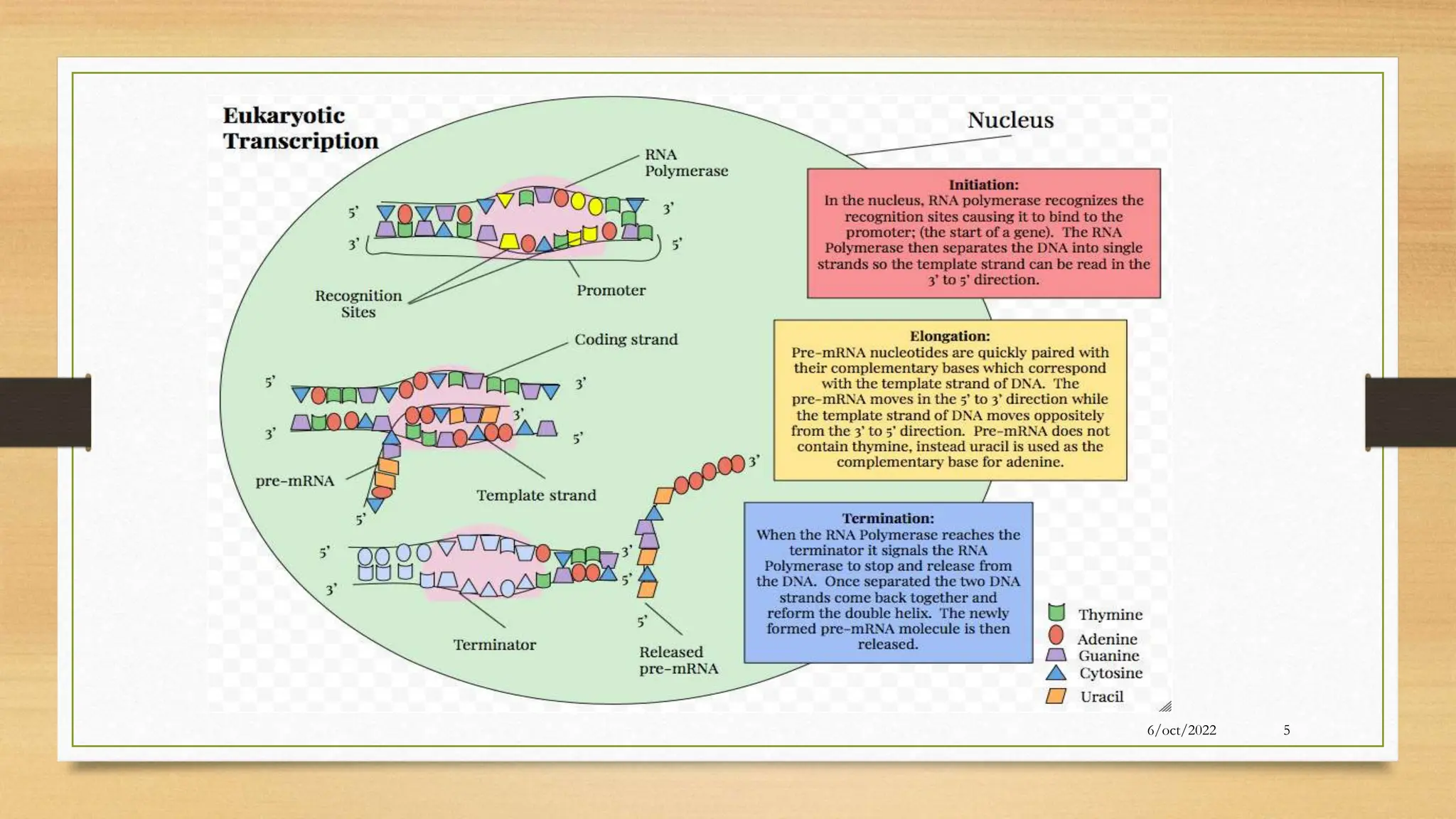
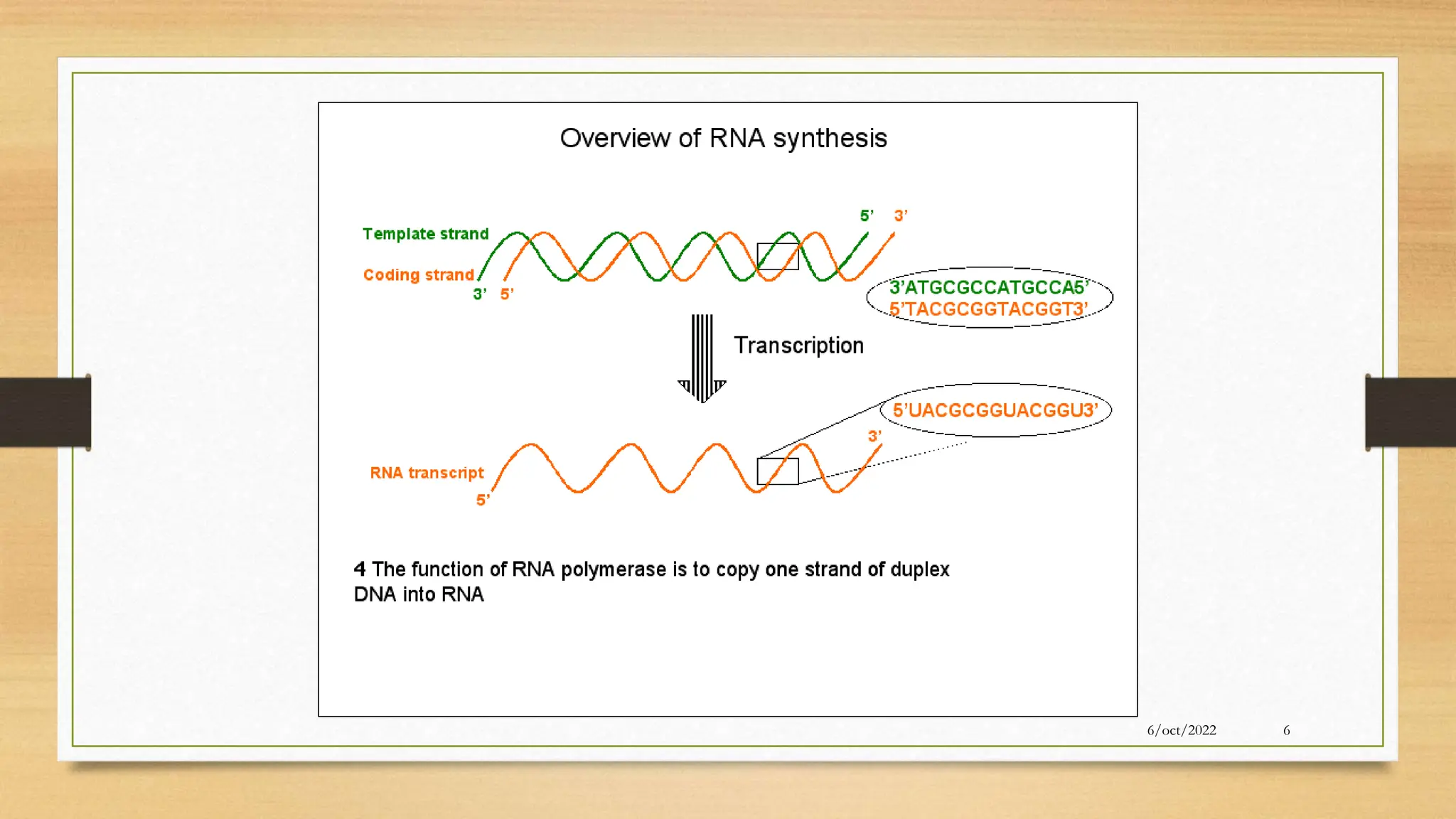
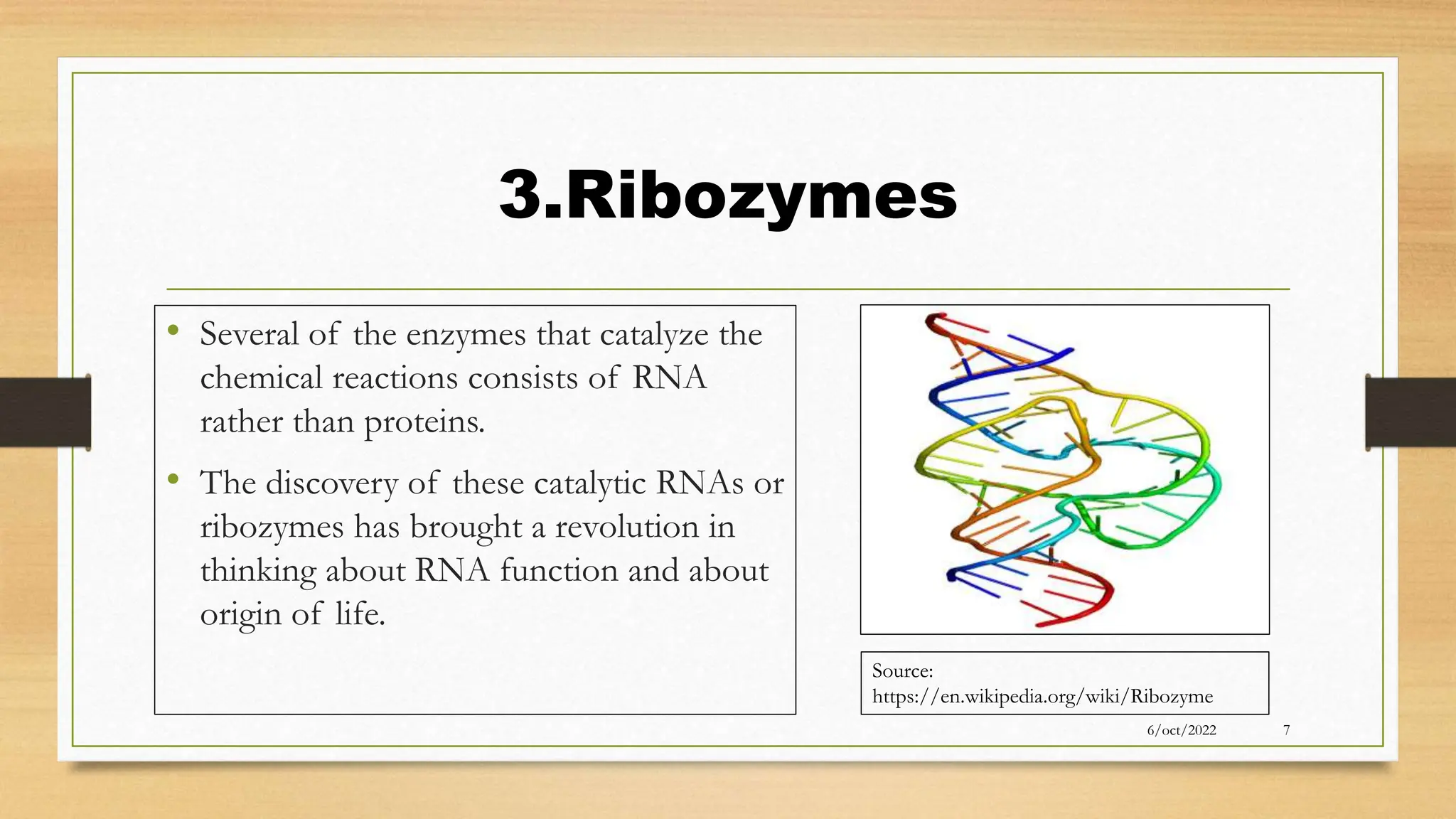
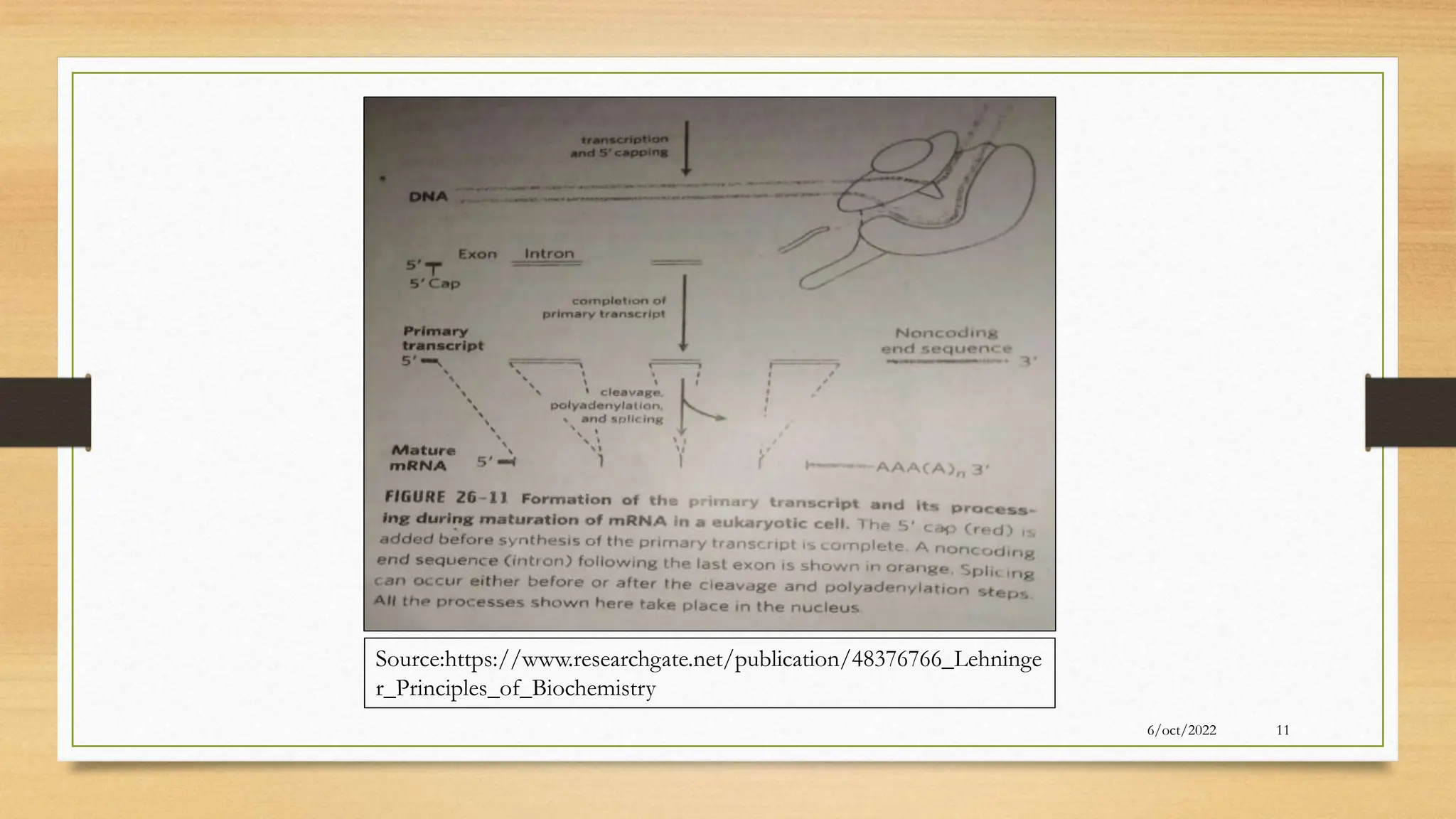
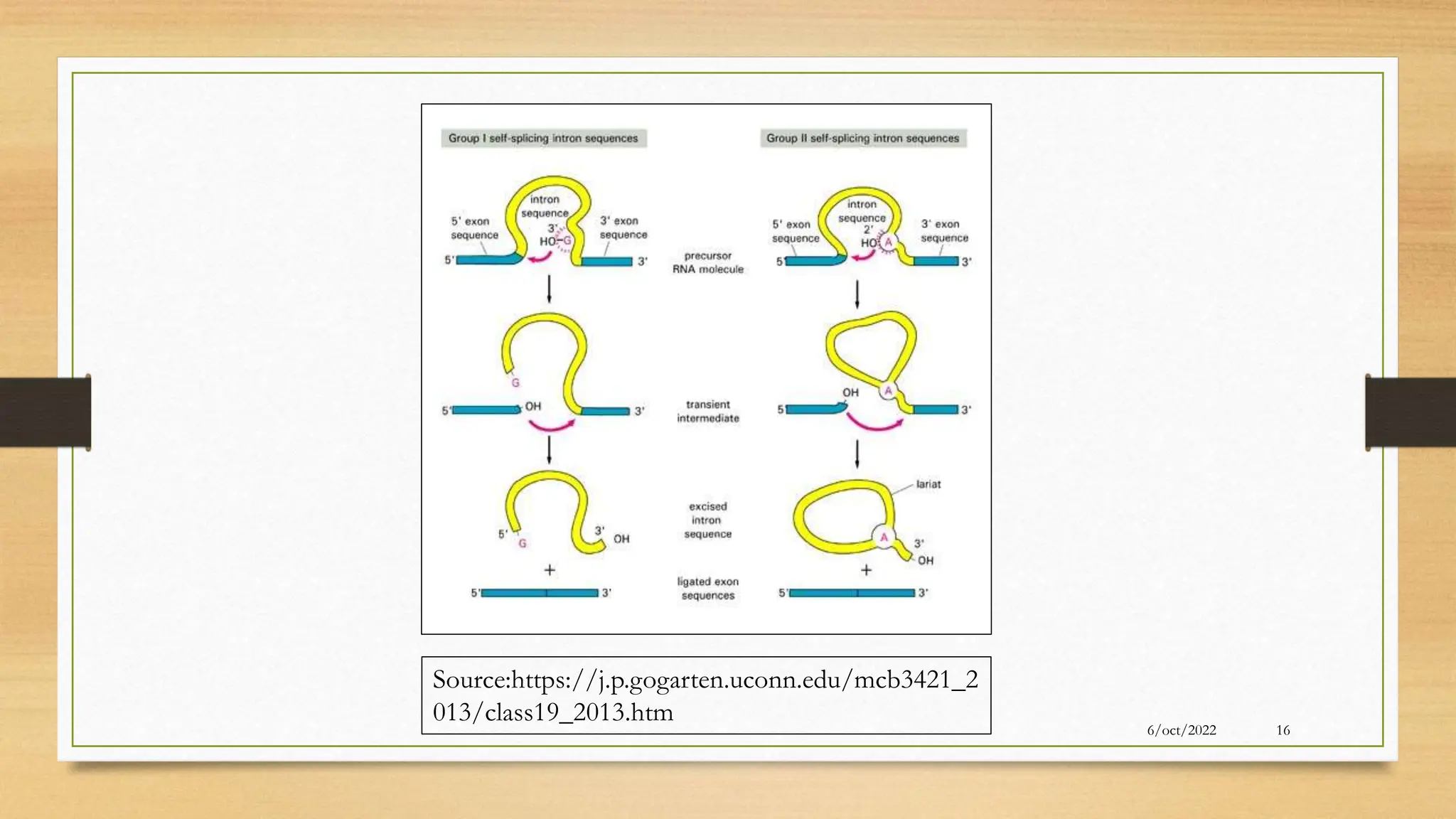
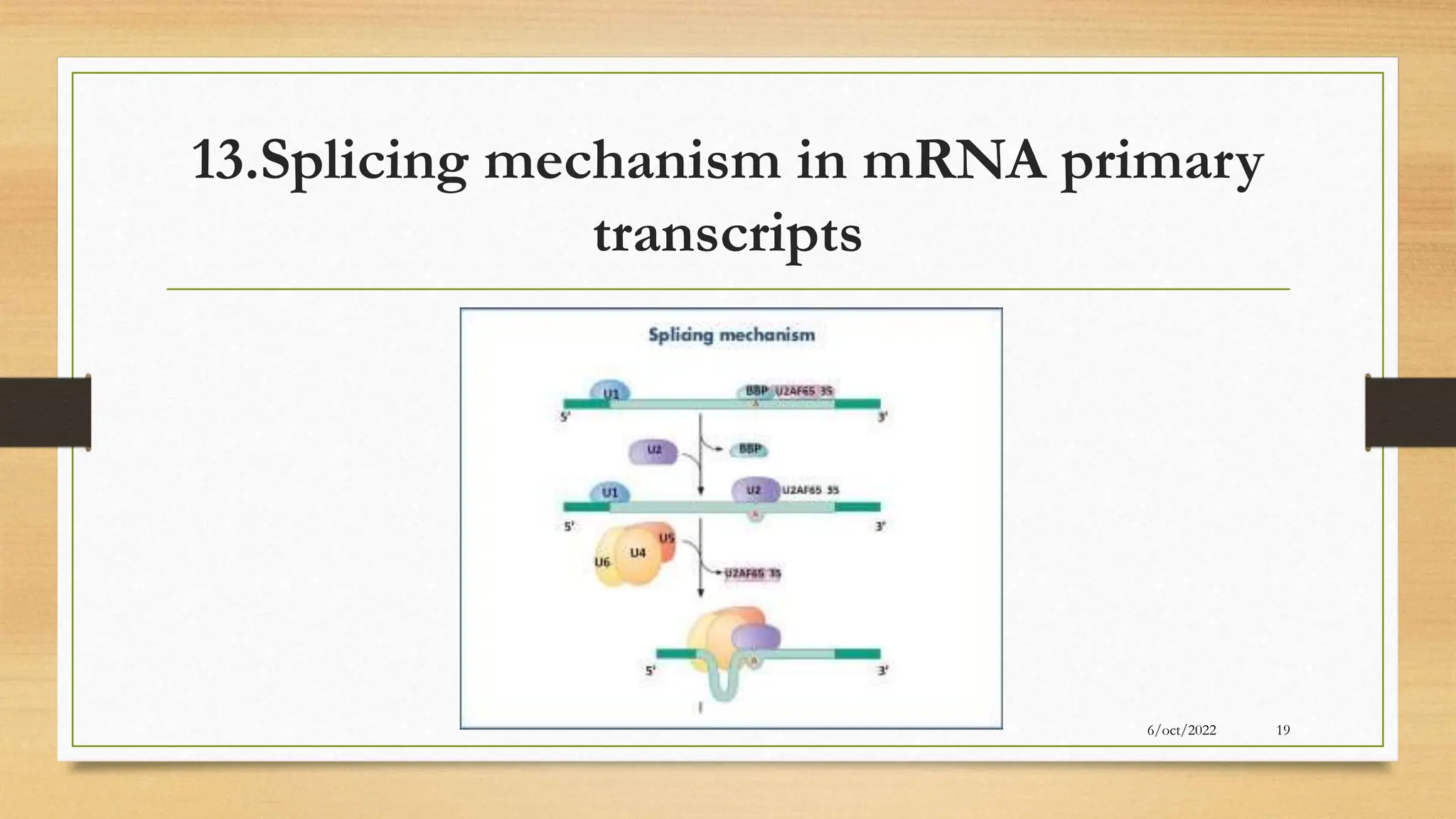
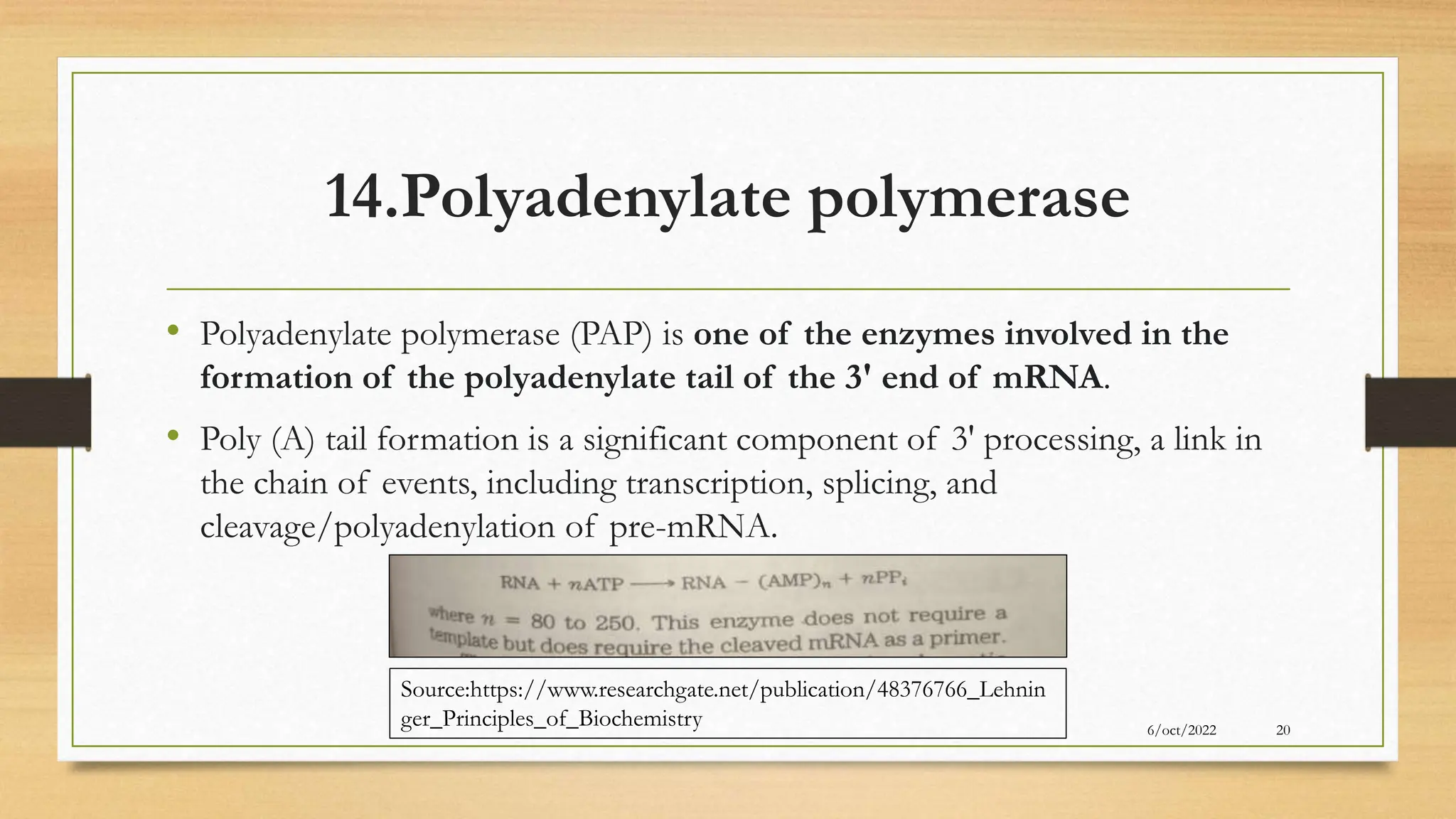
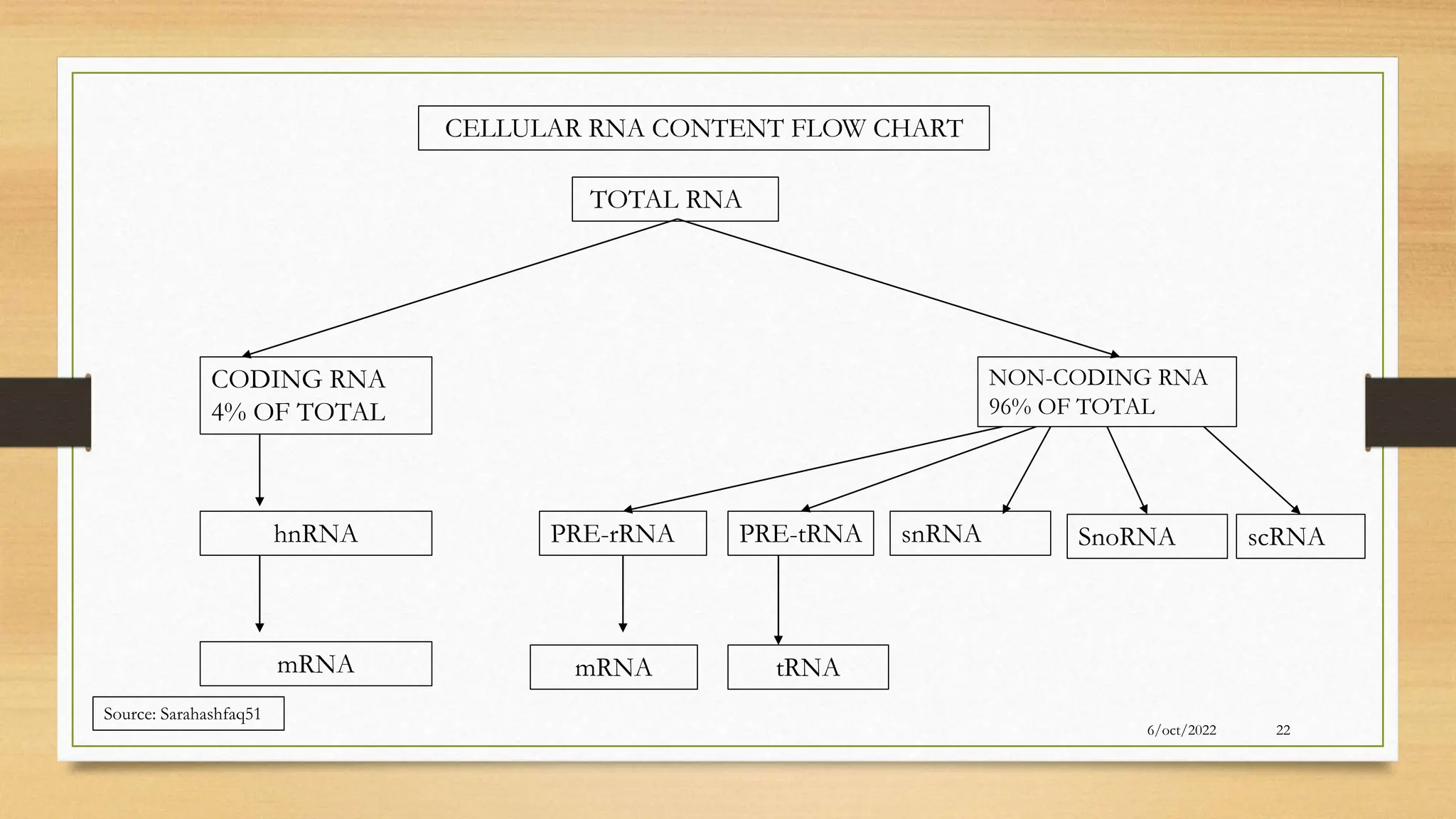
The document summarizes RNA synthesis and splicing. It discusses that in eukaryotes, RNA is processed after synthesis from DNA in the nucleus. There are three main types of RNA splicing - self-splicing via ribozymes, spliceosomal splicing involving protein complexes, and polyadenylation of mRNA. Introns are non-coding regions that are spliced out, while exons are coding regions that are joined to form mature mRNA. Small nuclear RNAs and proteins form spliceosomes that catalyze splicing of spliceosomal introns in eukaryotic pre-mRNA.