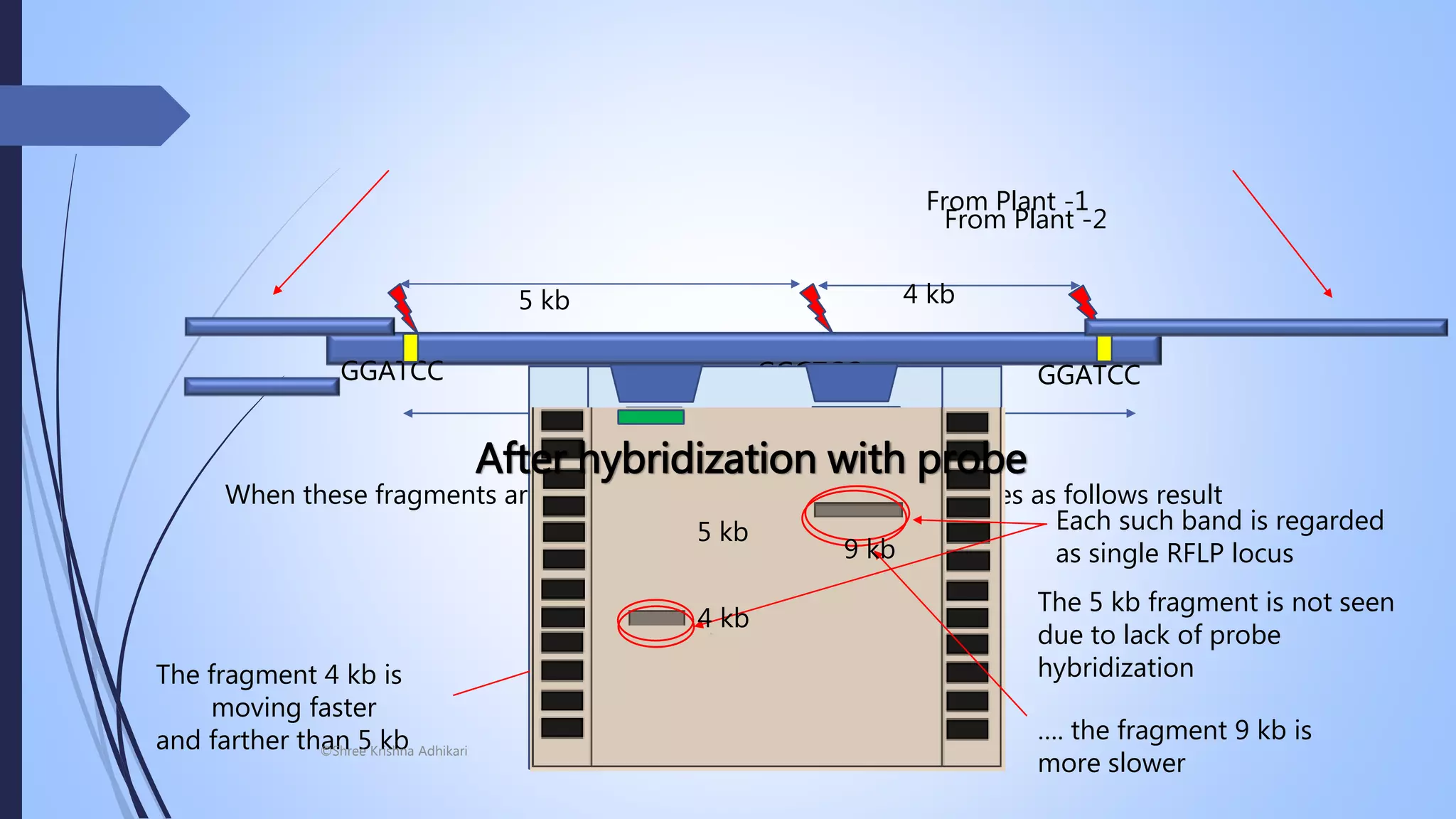
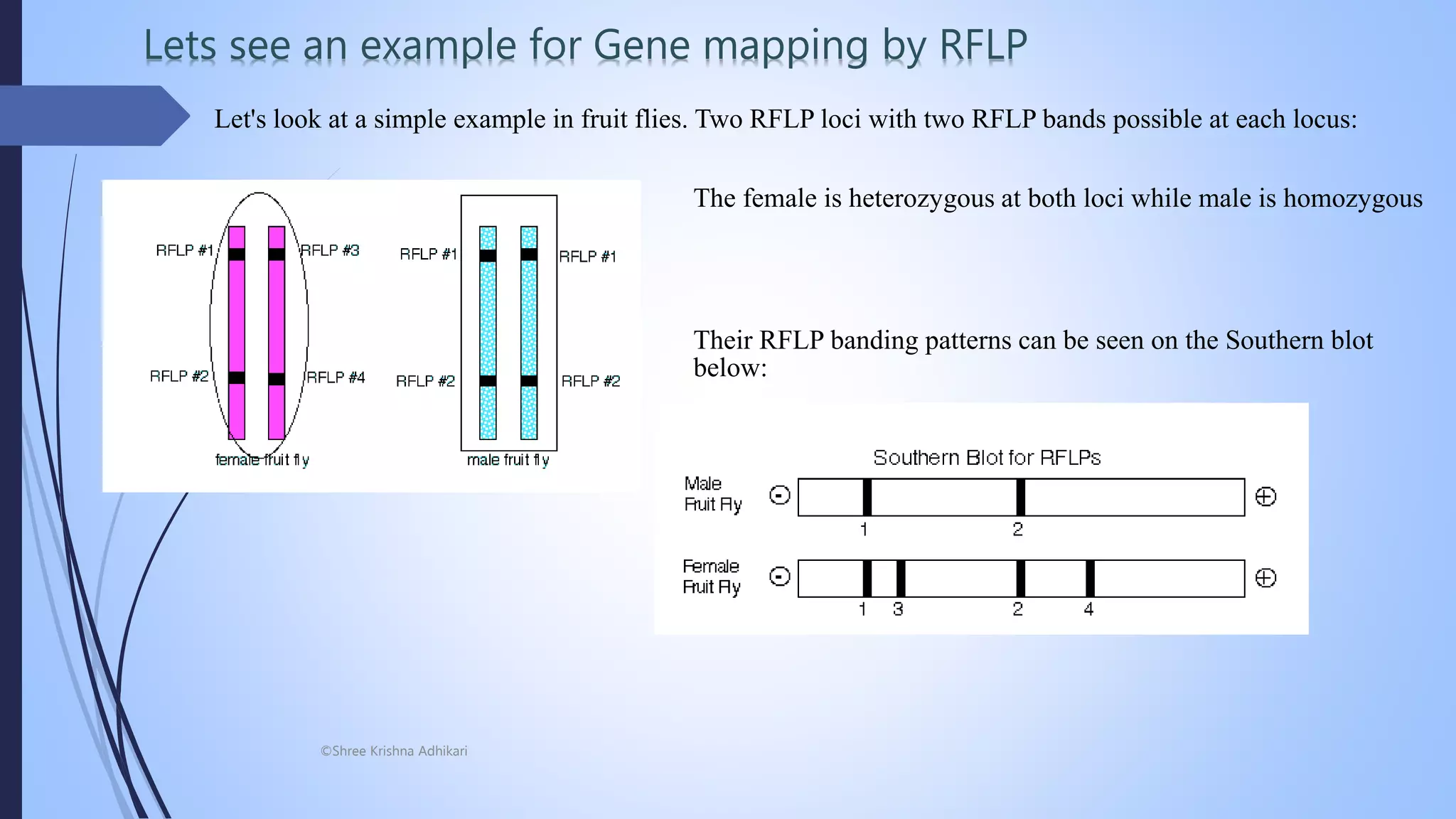
The document discusses plant breeding techniques focused on PCR-based and non-PCR based molecular markers, specifically restriction fragment length polymorphism (RFLP). It emphasizes the process of RFLP analysis, which highlights variations in DNA fragment lengths due to differences in restriction sites, aiding in genetic mapping and the detection of specific alleles. Furthermore, examples such as fruit fly genetics illustrate the application of RFLP in understanding inheritance patterns and genetic distances between loci.