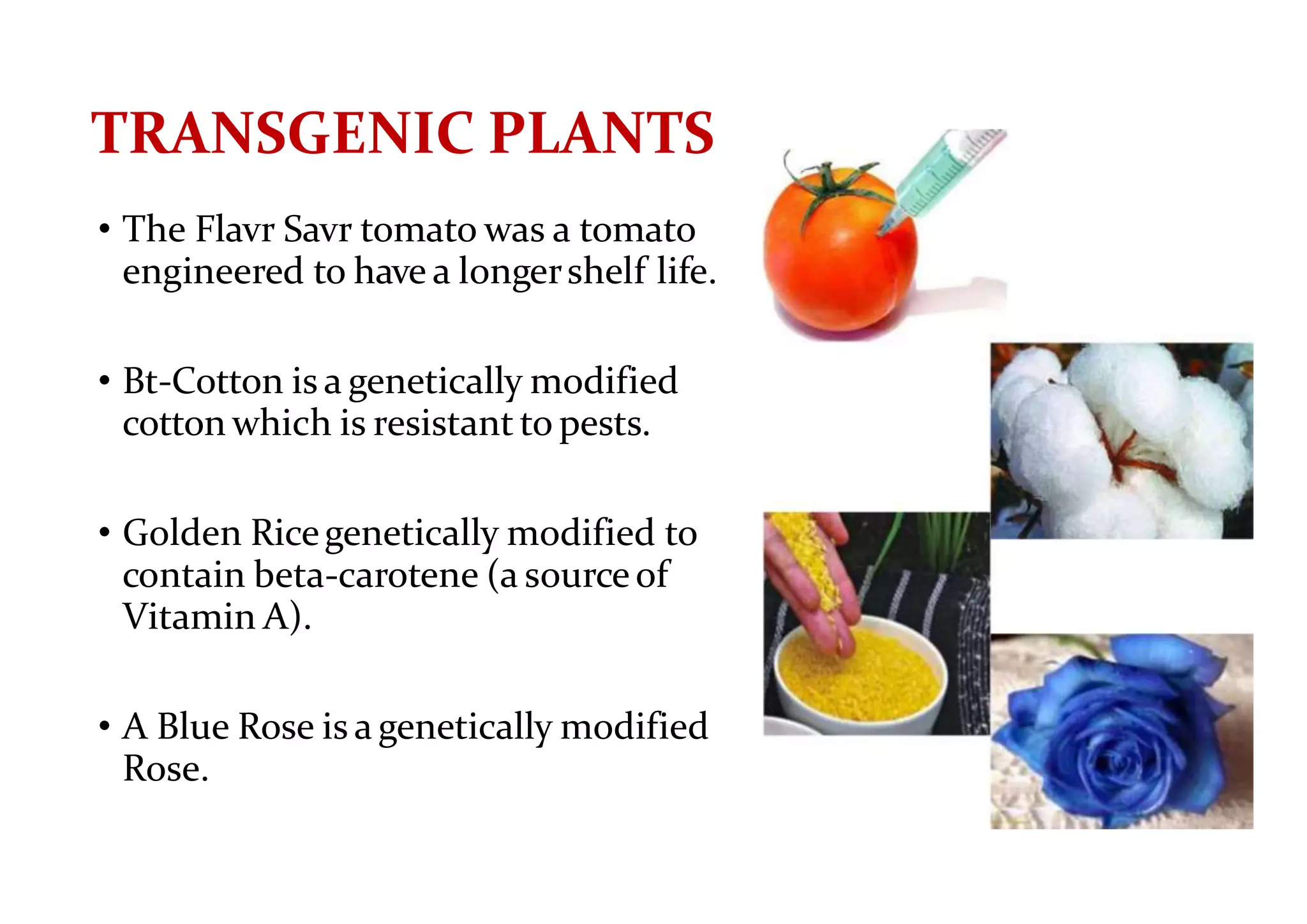
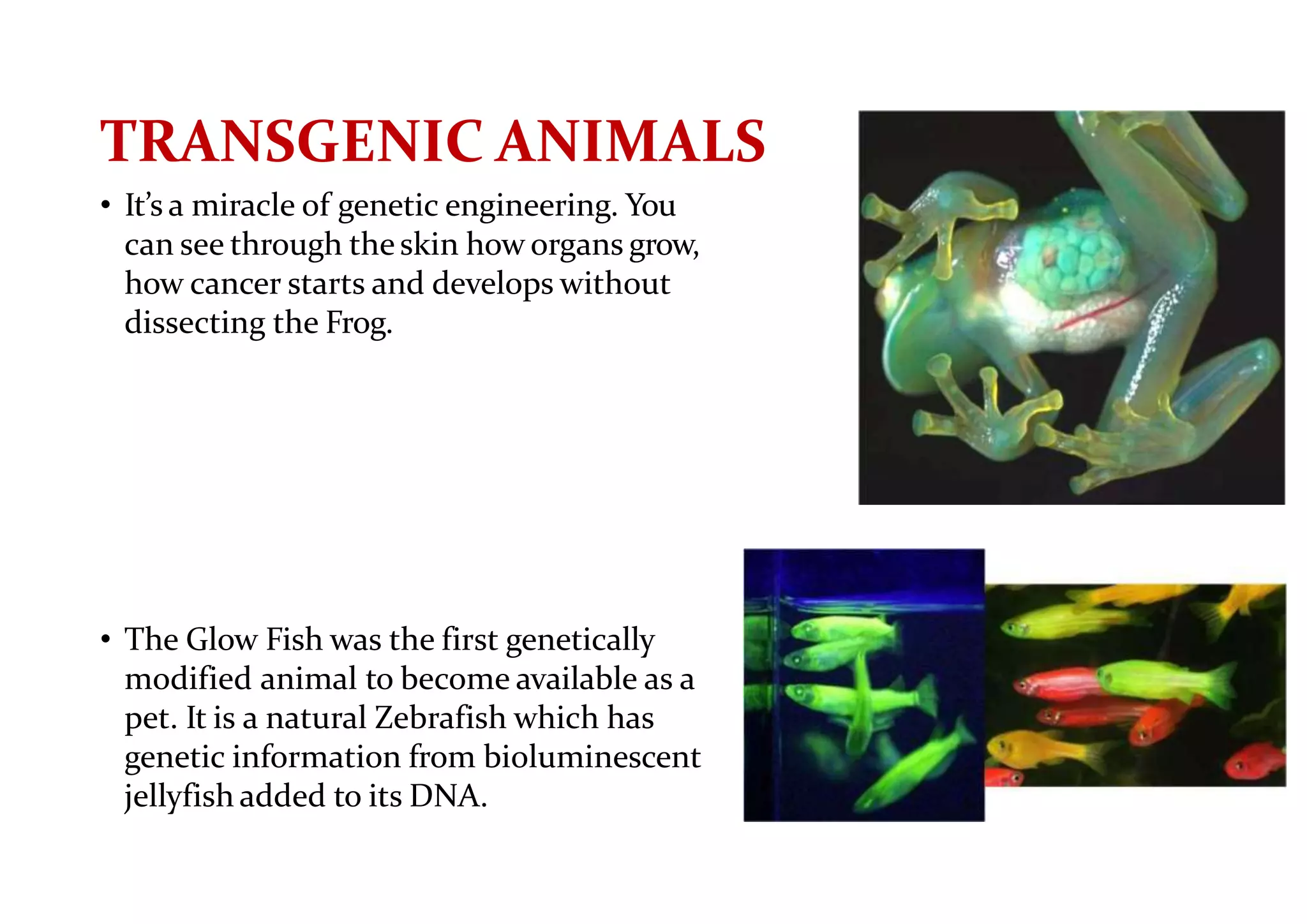
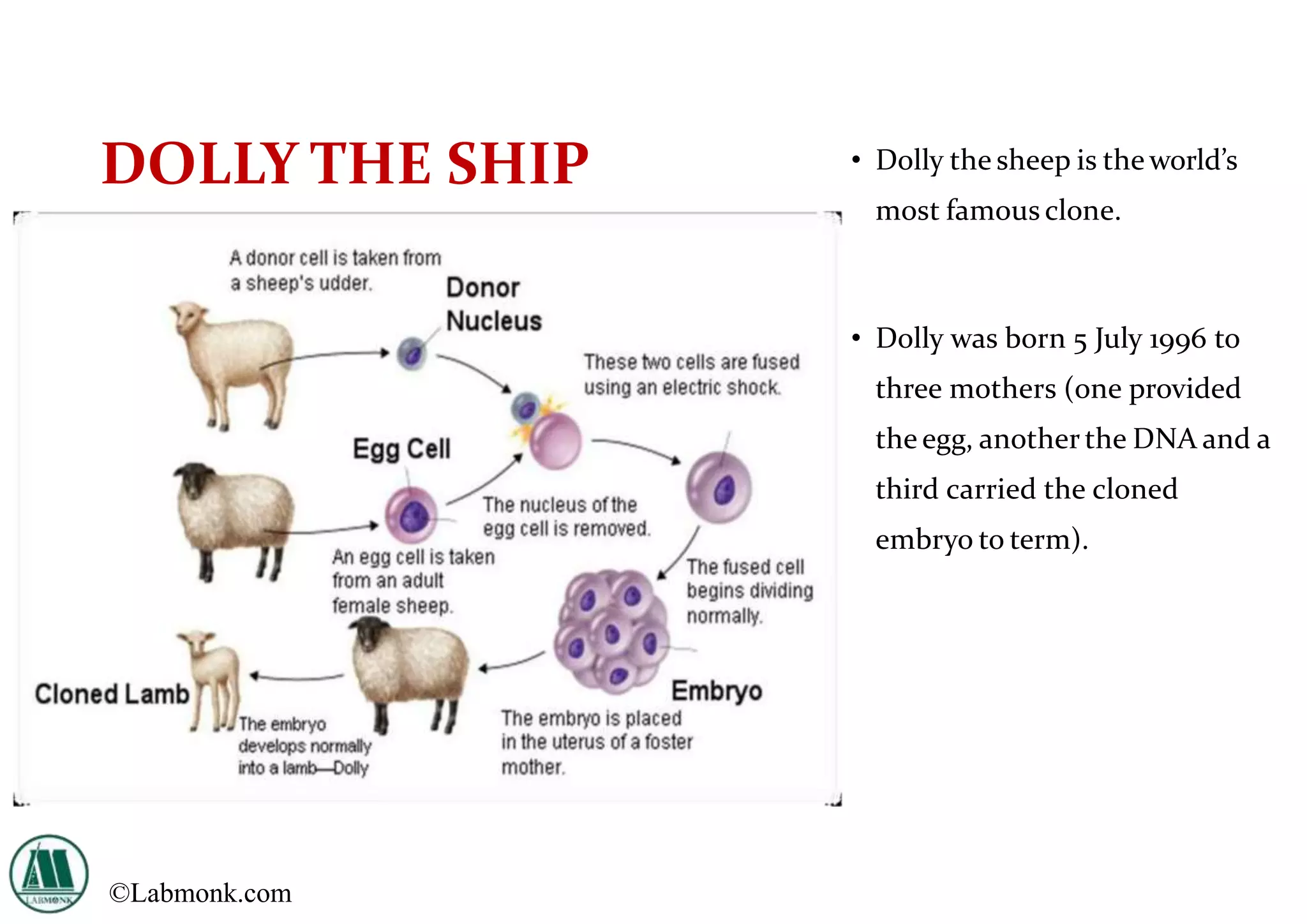
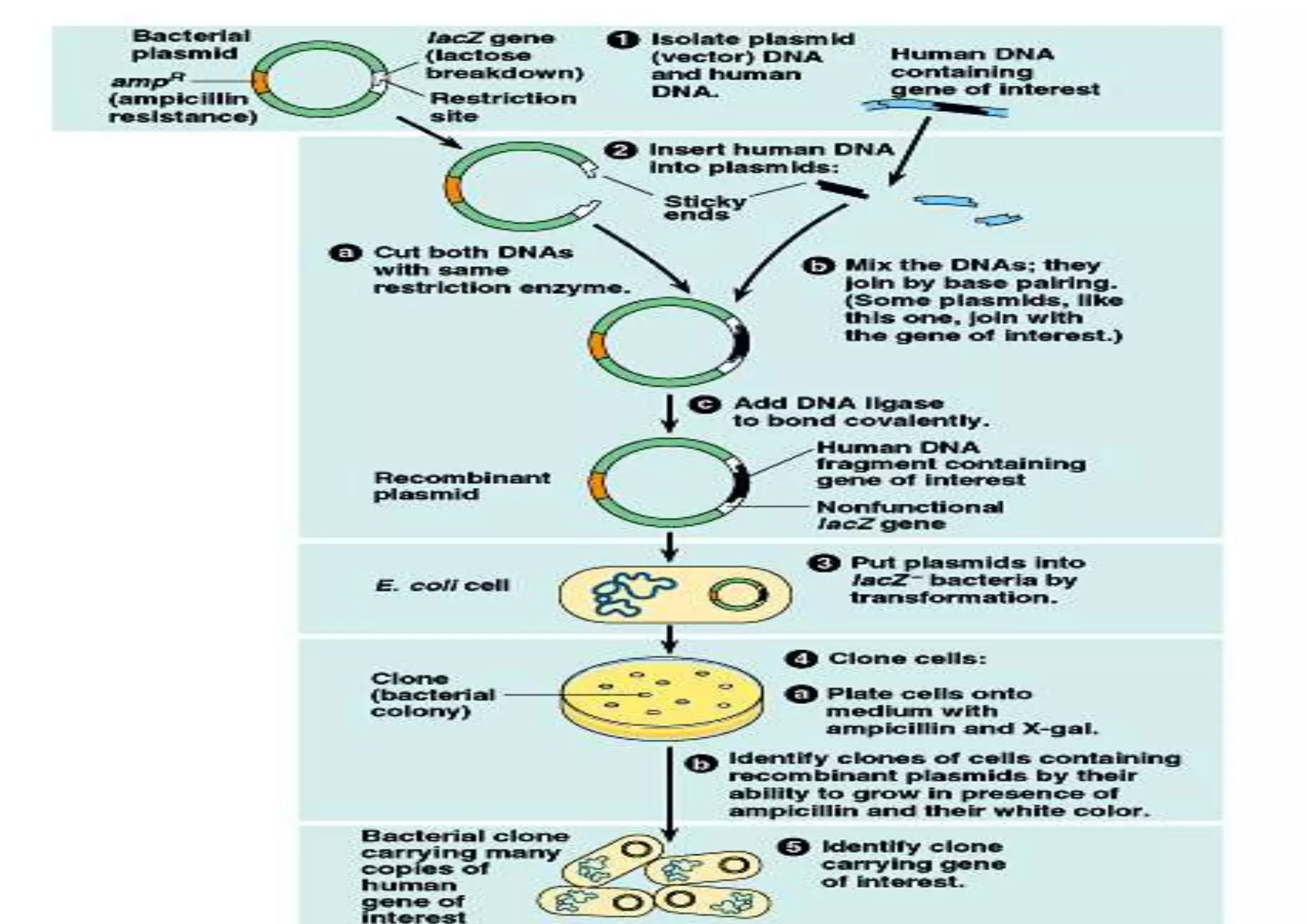
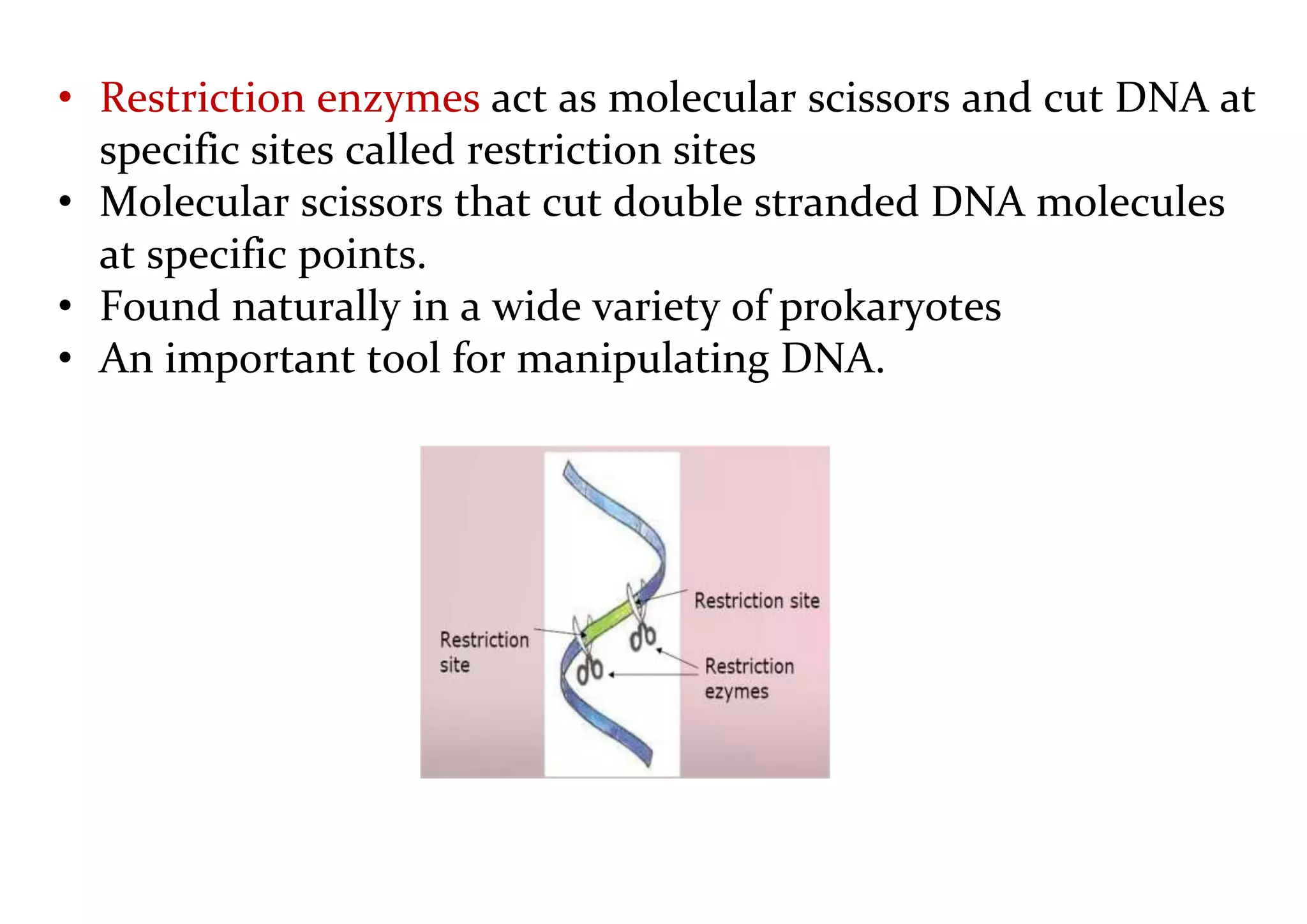
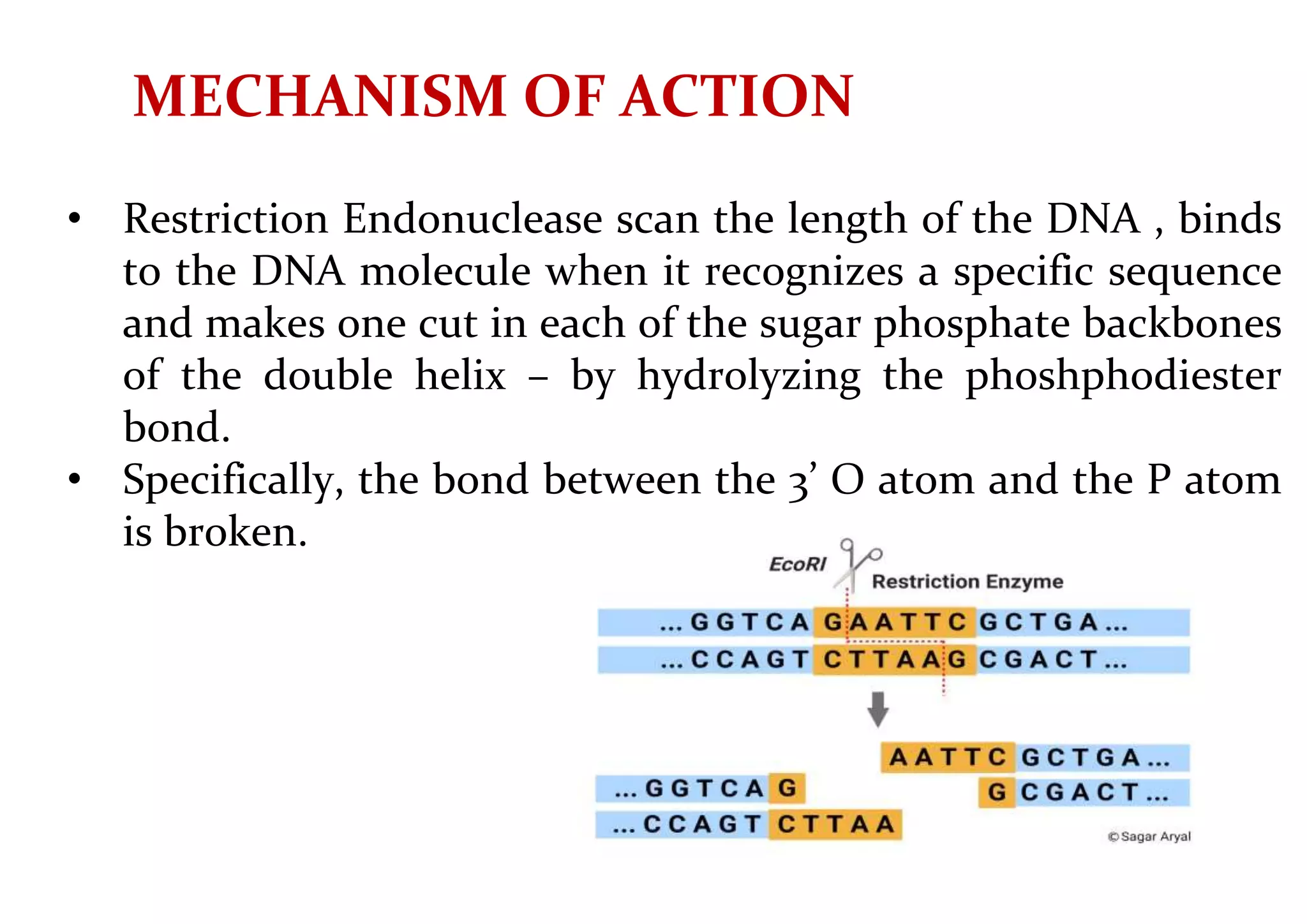
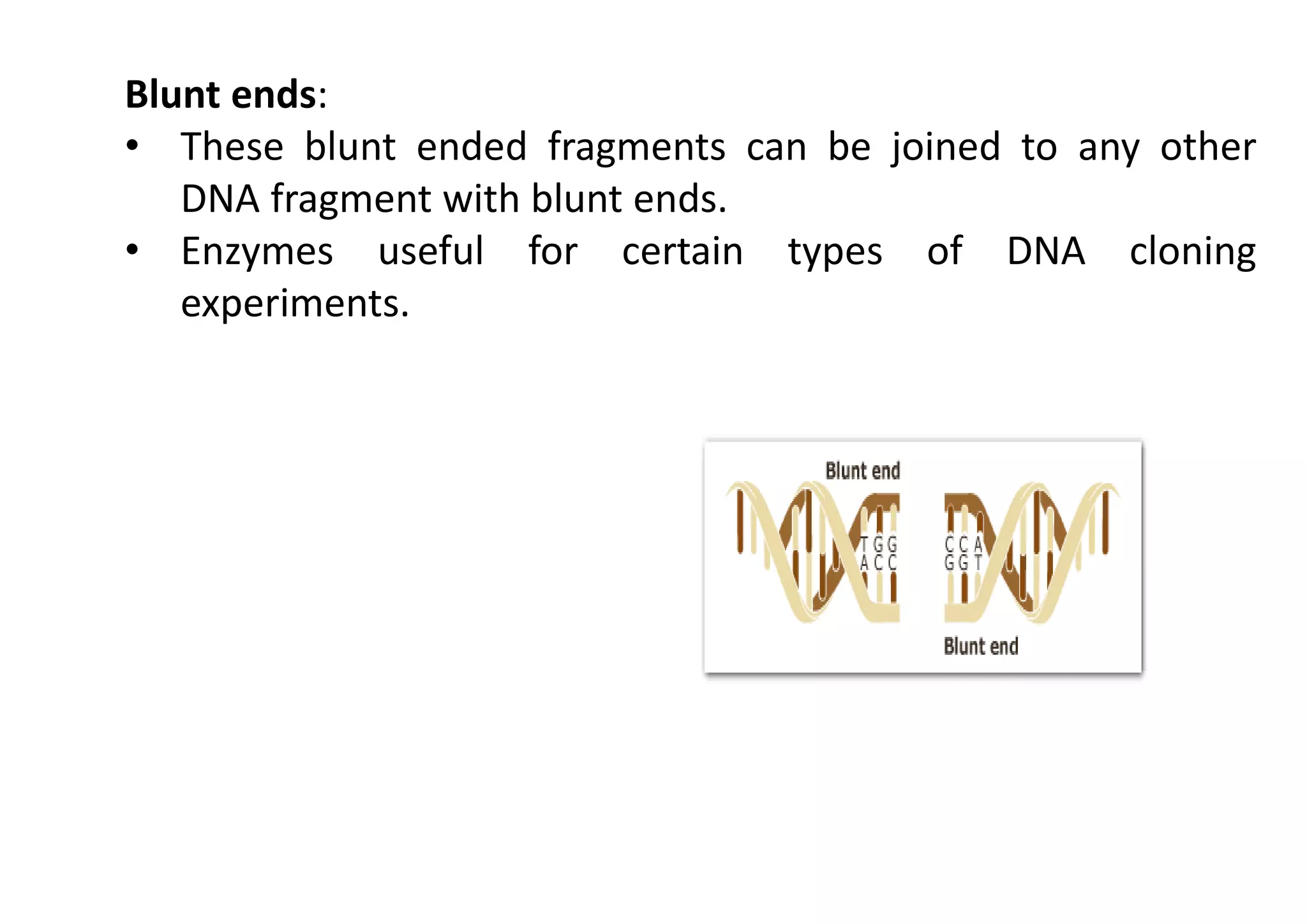
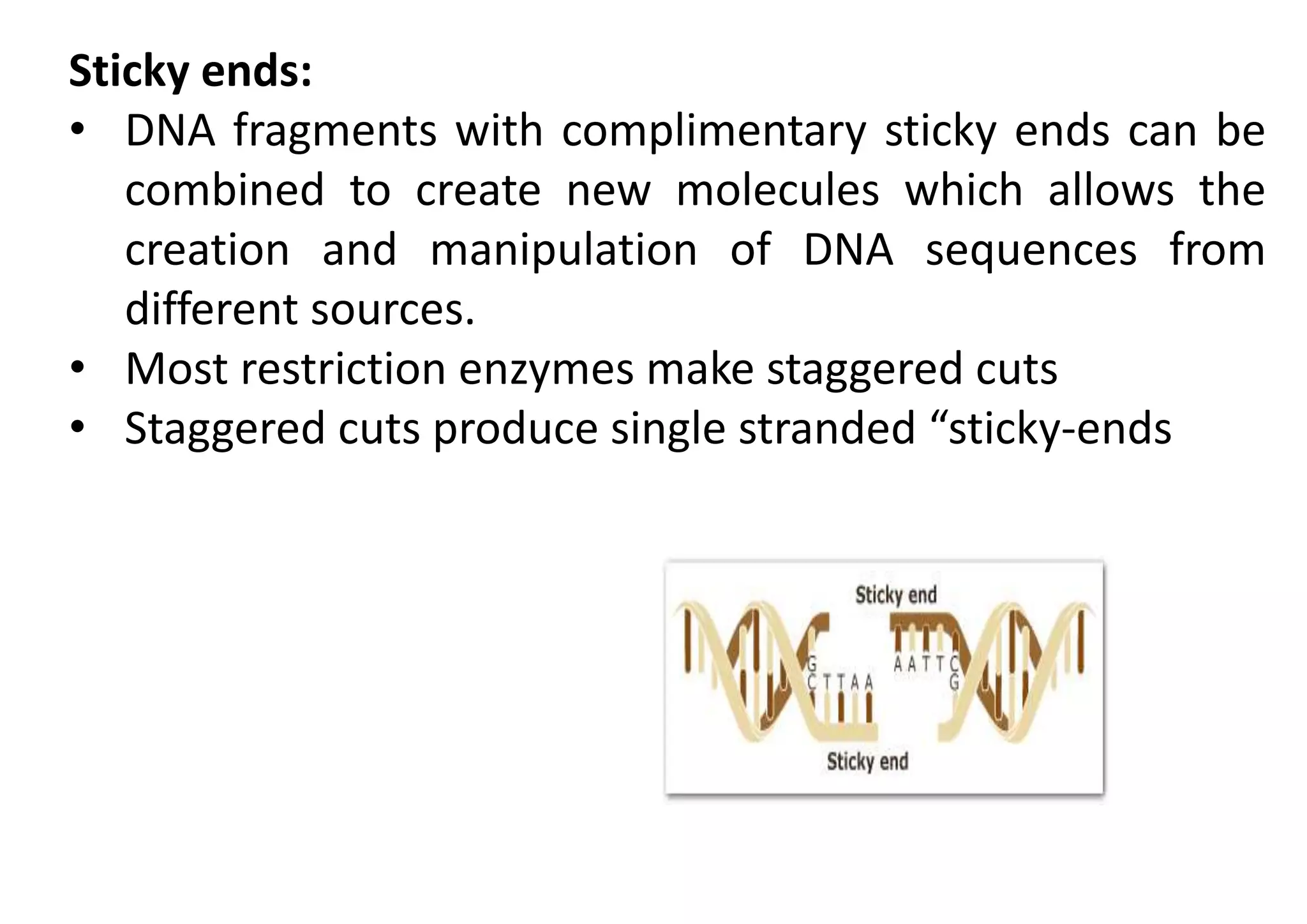
The document provides a comprehensive overview of genetic engineering, defining it as the manipulation of genes to produce desired traits in organisms, and discussing its historical development since the 1970s. It describes molecular tools like restriction enzymes and their applications in creating genetically modified organisms (GMOs) and transgenic animals, as well as processes such as insulin production. Additionally, the document outlines the mechanisms of action for various types of restriction enzymes and their significance in biotechnology.