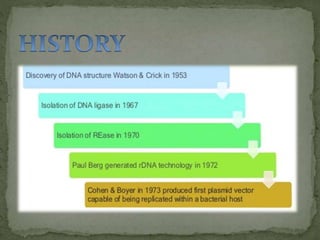
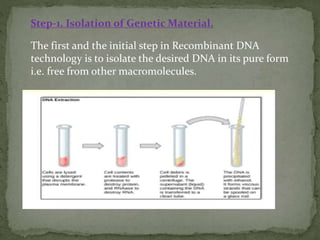
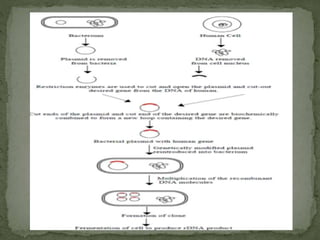
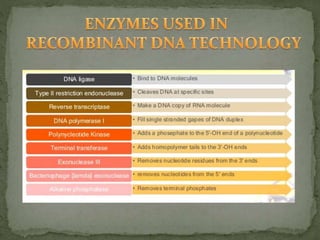
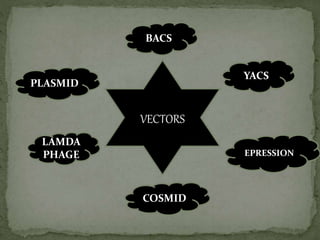
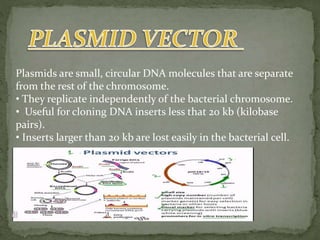
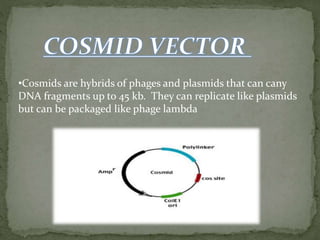
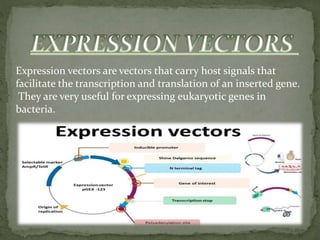
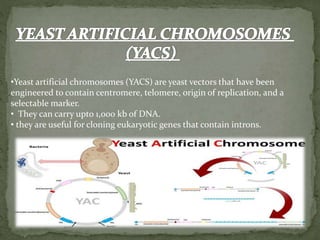
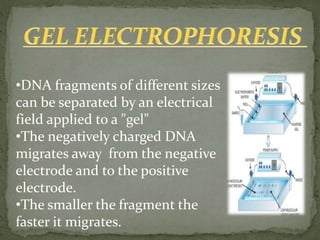
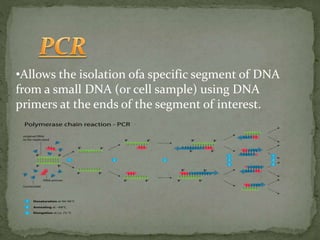
Recombinant DNA technology, also known as genetic engineering, involves combining DNA from different sources to create new genetic sequences. The process includes key steps such as isolation of genetic material, cutting DNA with restriction enzymes, amplifying genes using PCR, ligating DNA molecules, and inserting recombinant DNA into host cells. Applications of this technology span agricultural modifications, medical therapies, and advancements in clinical diagnostics.