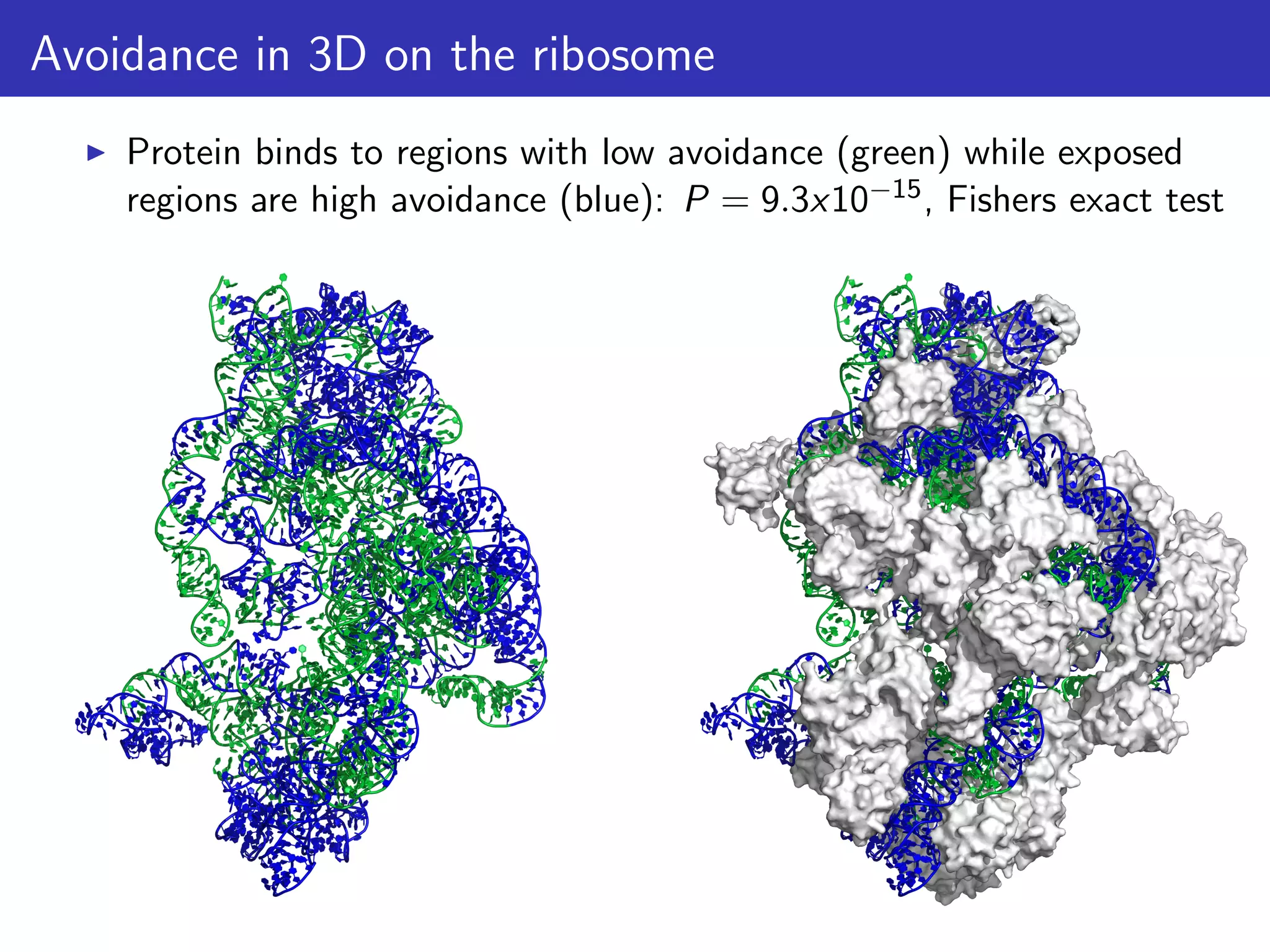
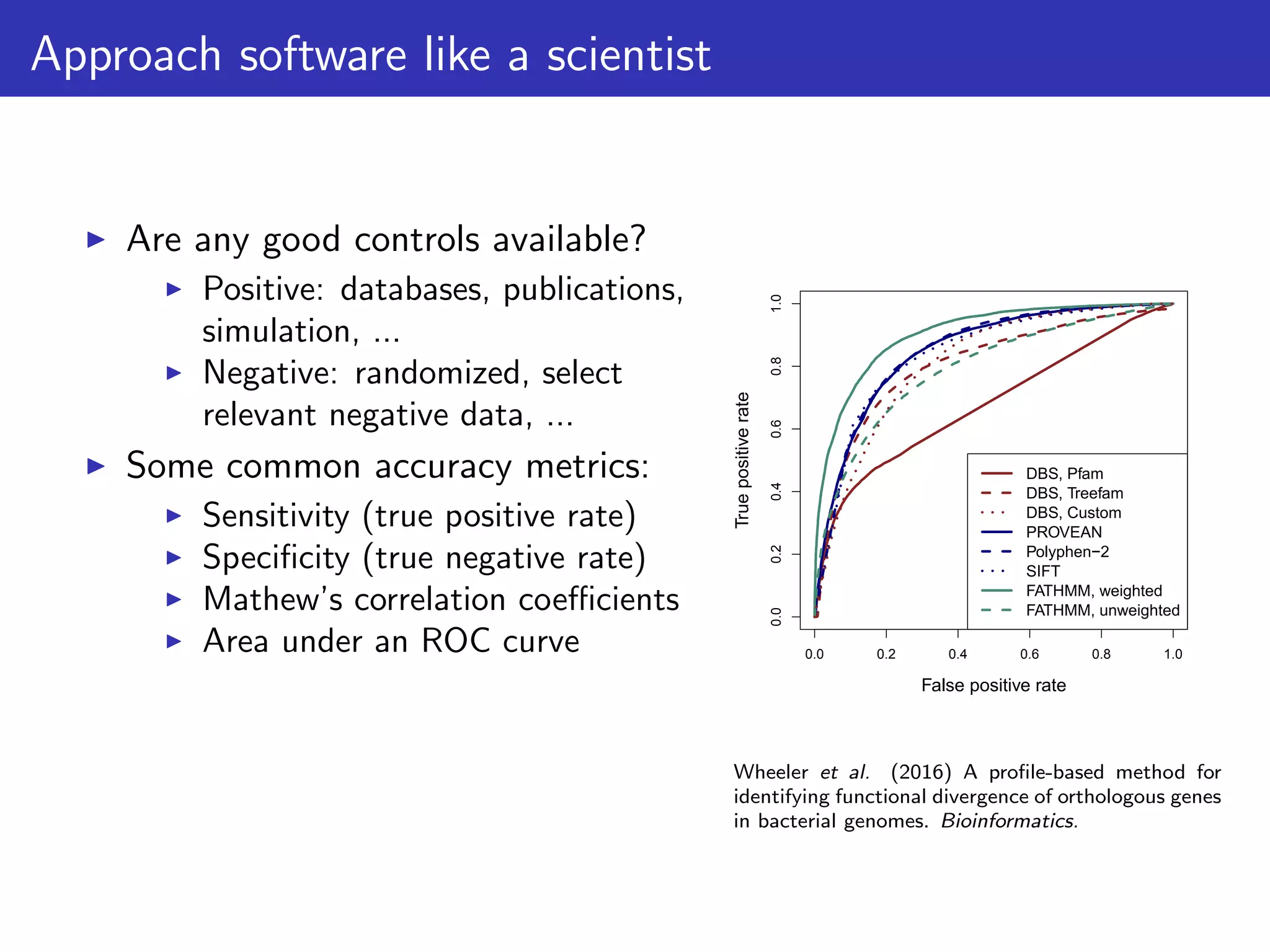
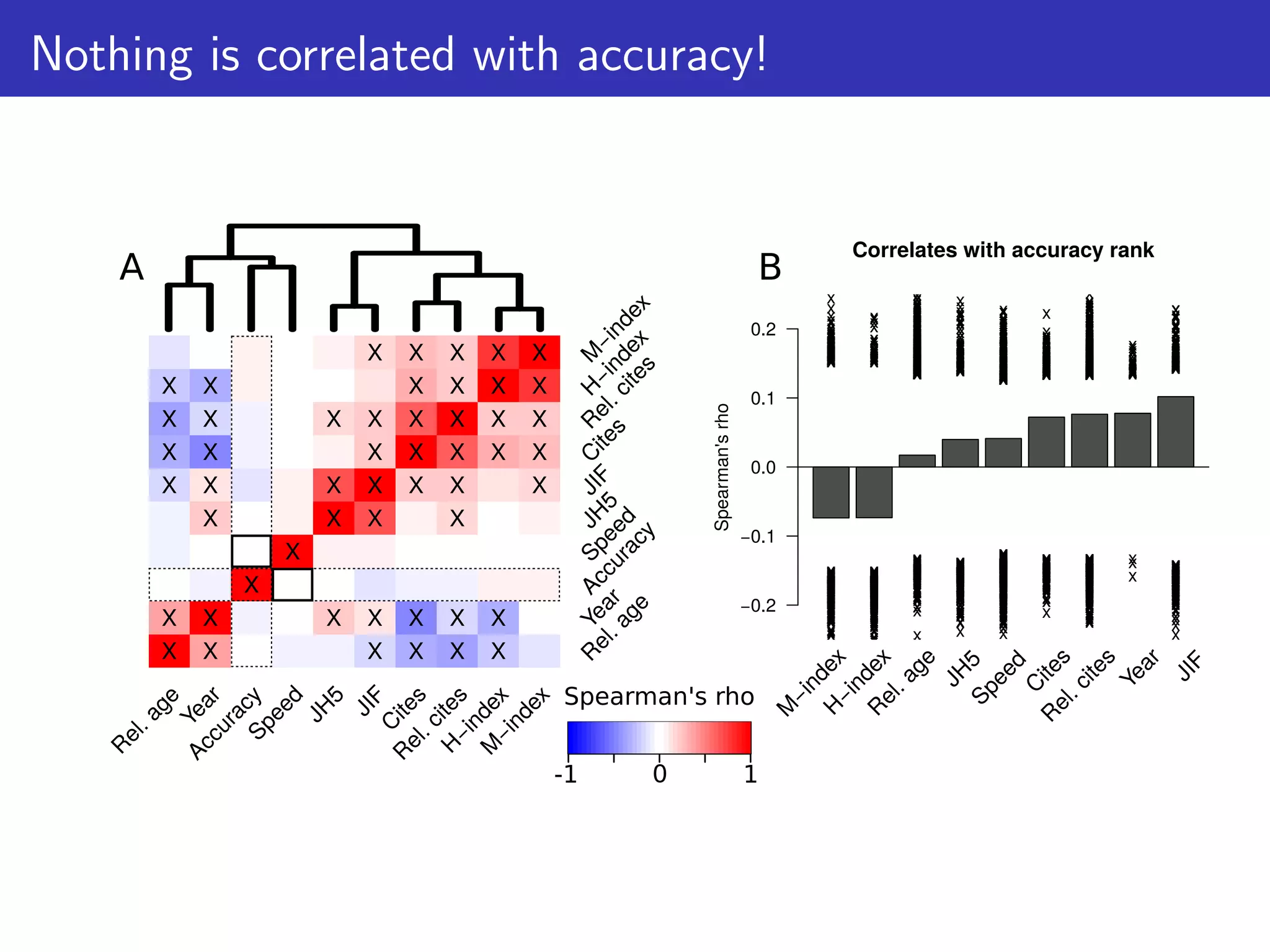
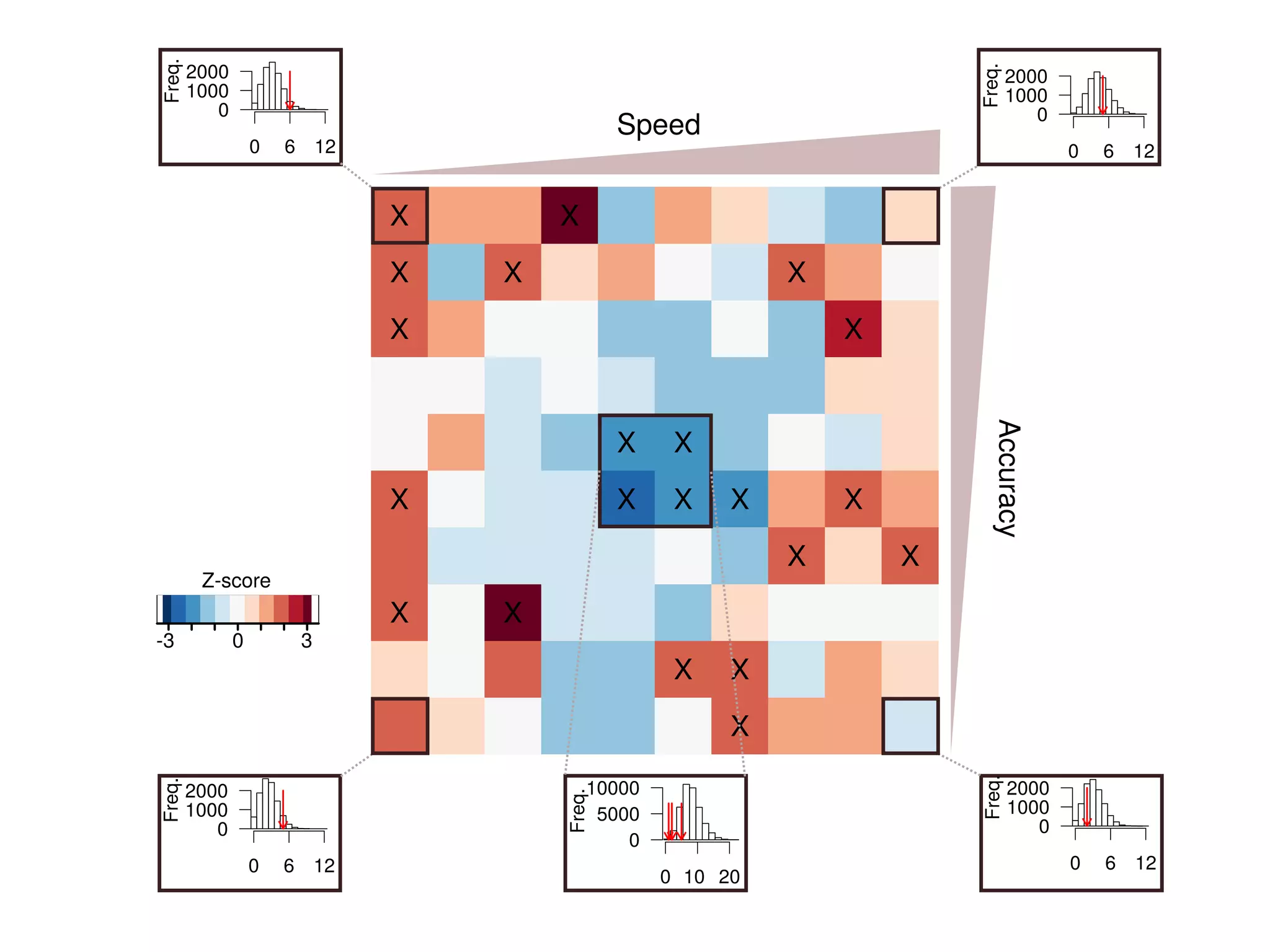
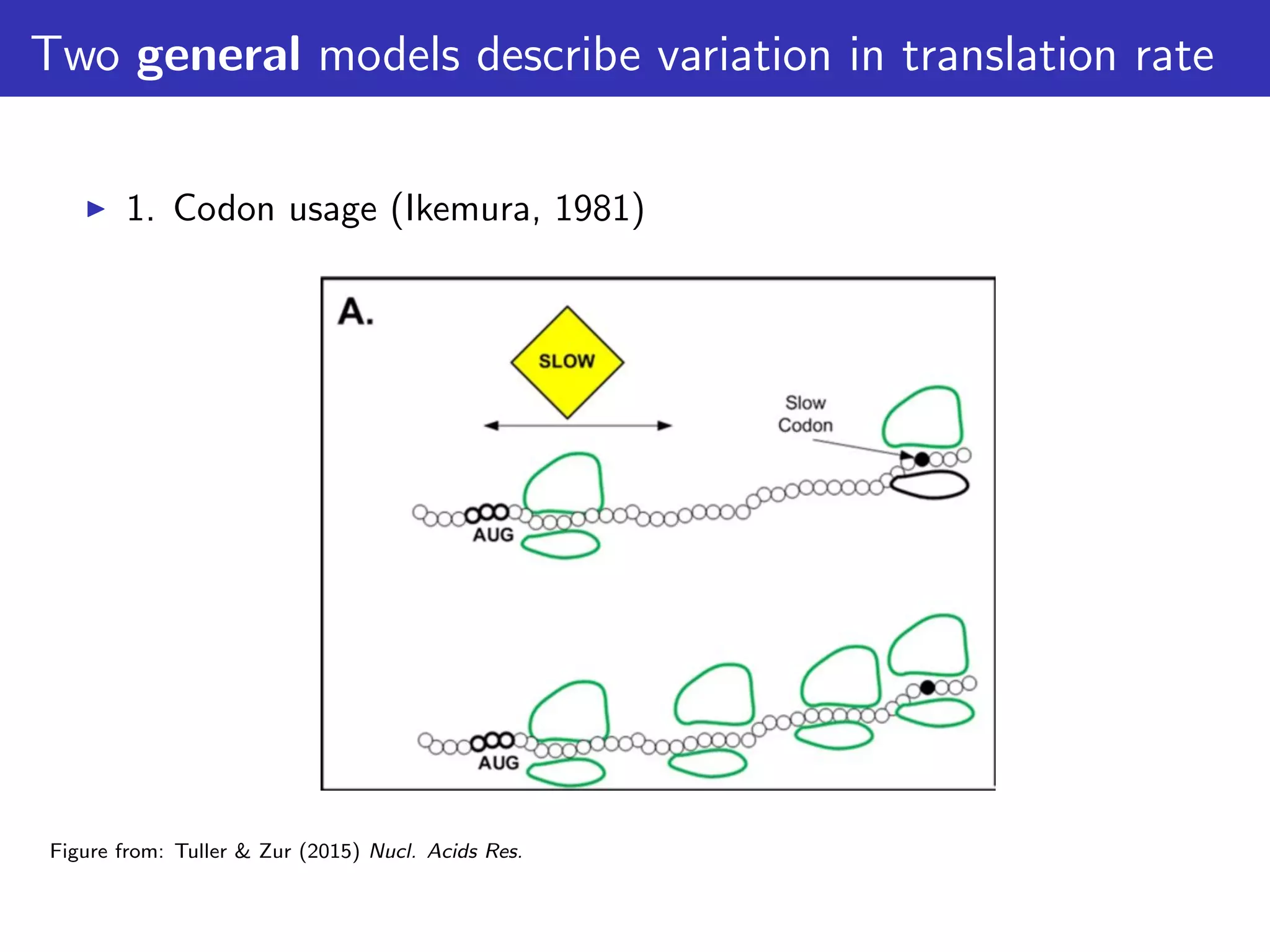
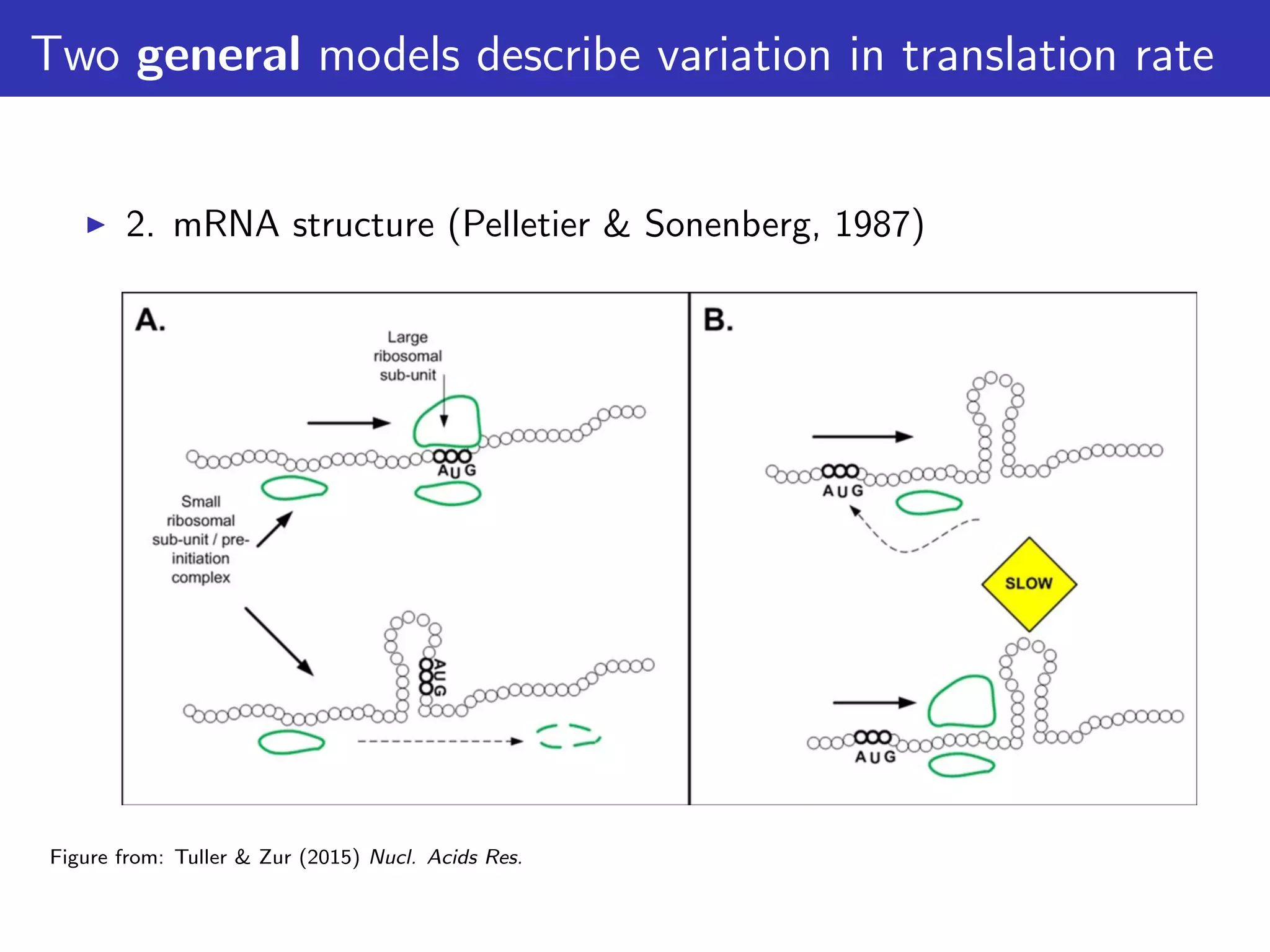
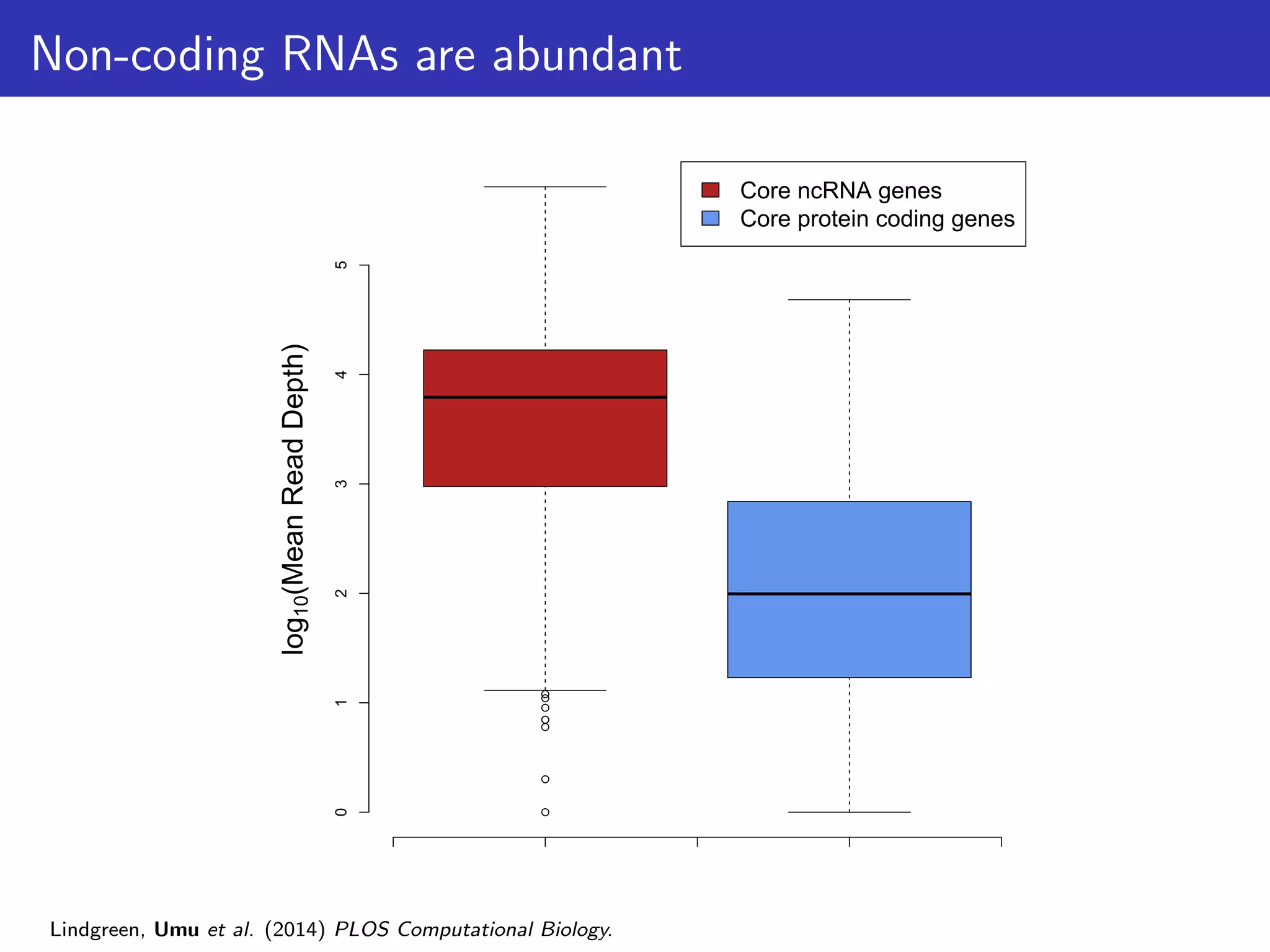
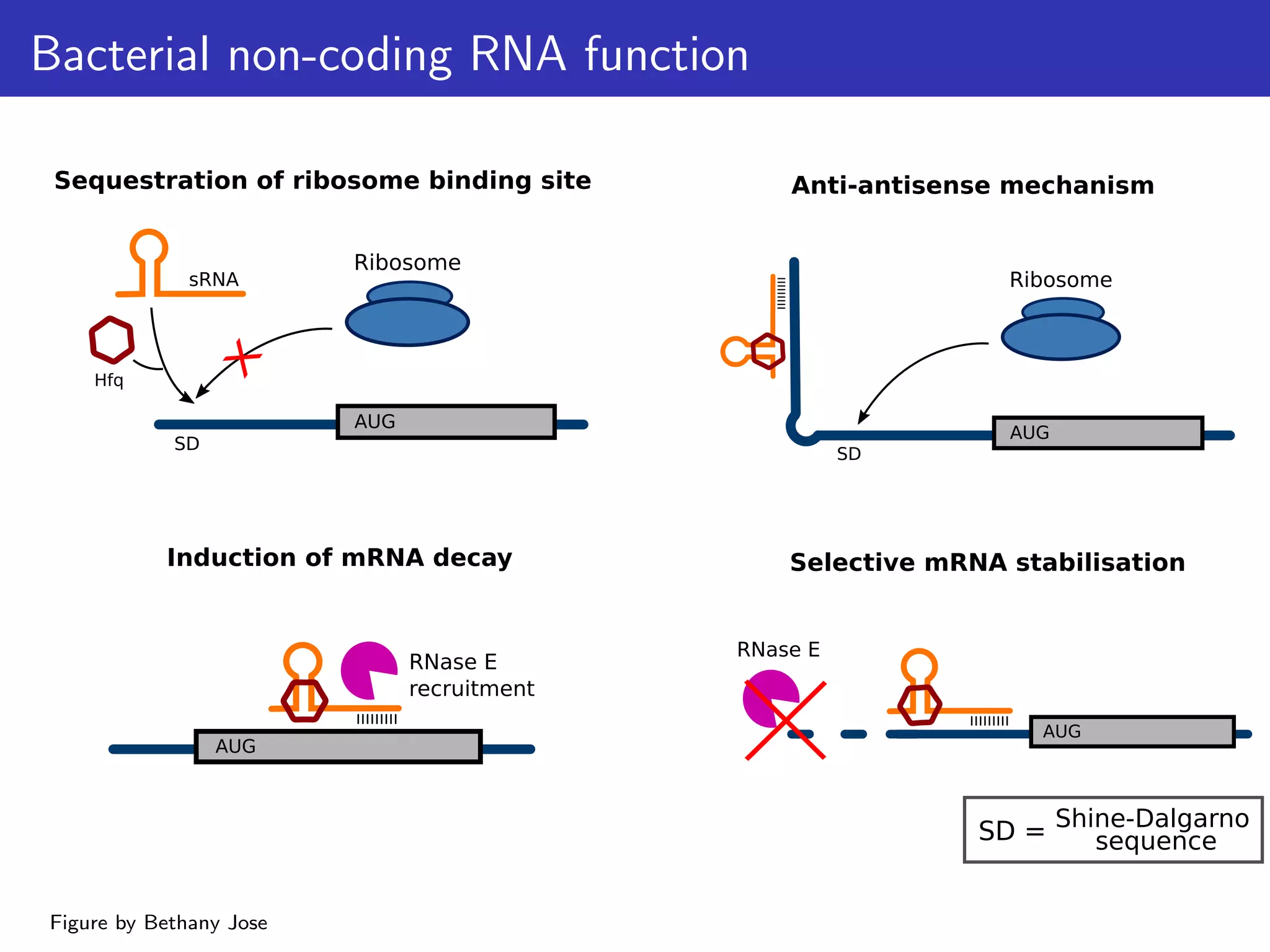
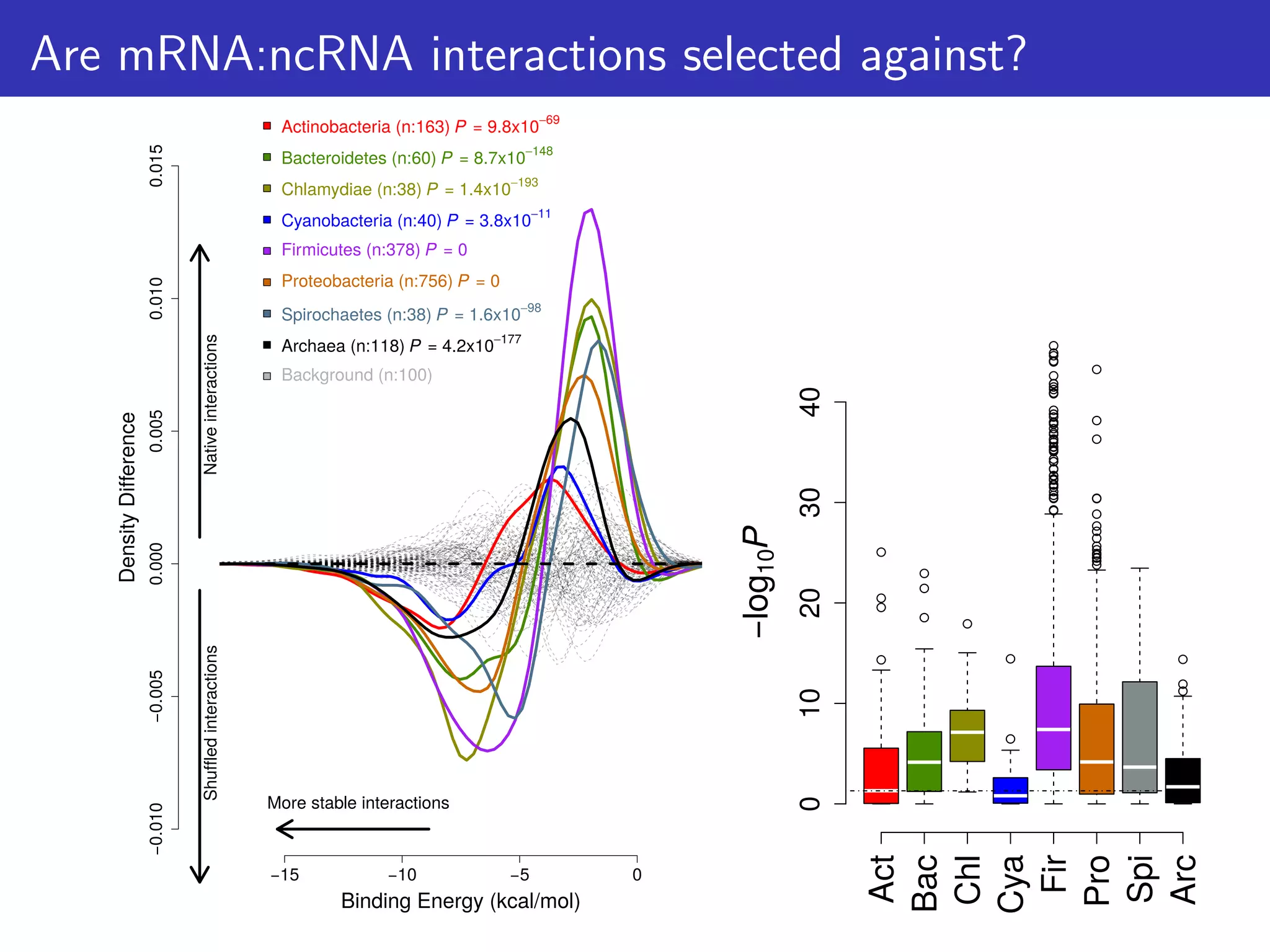
The document discusses the influence of non-coding RNAs (ncRNAs) on protein expression in prokaryotes, highlighting that mRNA levels do not always correlate with protein levels. It presents a hypothesis of selection against mRNA:ncRNA interactions and suggests testing it through a range of genomic data. Additionally, it addresses the challenges in bioinformatics related to software in the analysis of RNA interactions and benchmarking methods.

![Determinants of protein concentration
Protein concentration depends on mRNA concentration, translation and
degradation rates
DNA
[D]
RNA
[R]
Protein
[P]
ktranscription ktranslation
kmRNA degradation kprotein degradation
0 1
A
T GGC
TA
A
GGGGCA
A
T
C
T
T
TA
C
A A
G
AT
CC
G
T
T
C
C
T
G
A
AC
G
C
AC
T G
C
G
T C
G
G
G
A
A
C
G
T
G
T
T C
CAGTTTCTATTTATT
T
G G T G A A T G GTATTA A G C T GC
AA
G
G G
C
AA
A
T
C
G
A
G
T
C
T
TT
T
G
A
T
C
AG
T
T
C
G
T
G
A
T
C
C
T
G
T
T
G
A A
A
A
A
C
A
C
G
G
T
C
A GC
C
A
G
A
T
G
G
T TT
A
C
A
A
GC
A
C
G
C
G
A
T
T
T C T A
C
T
G
T
T G T C C CG
T CT
C
G C C C G G T T T C
T
C
AT
CA
CA
GTAA
CAACGCCG
GT
GGC
G
G
T
A
C
C
A
G
C
A
G
T
A
A
C T A C C A T
C
A
TGGTAGCAGCG
C
G
C A
G
A A
T
AC
T
T
CC
G
C
G
C
A
ACAGG
A
C
A
G
C
G
A
A
GAAACCG
A
A
TAA
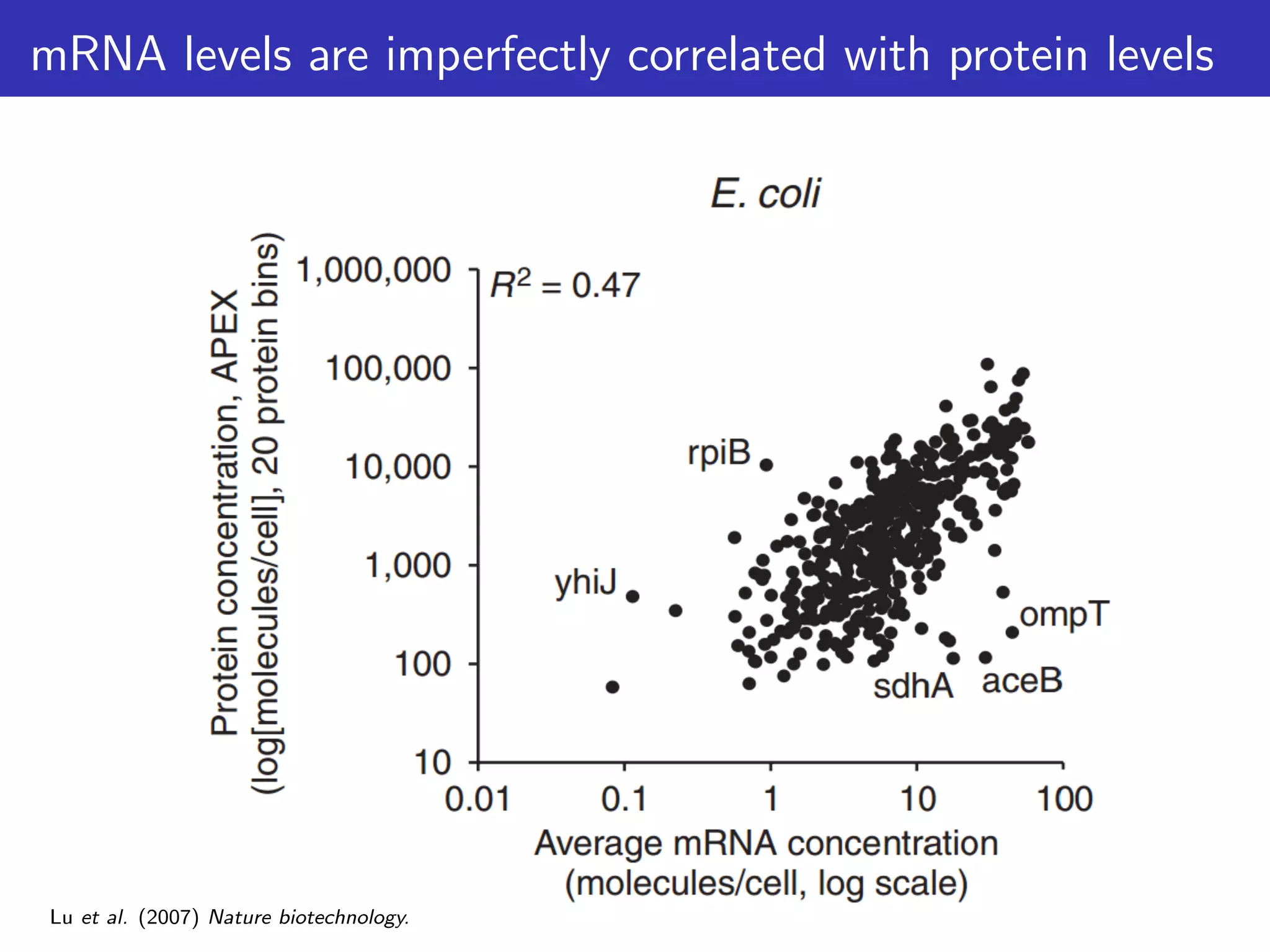
de Sousa Abreu, Penalva, Marcotte & Vogel (2009) Global signatures of protein and mRNA expression levels. Molecular
BioSystems.](https://image.slidesharecdn.com/talk-161115083940/75/Random-RNA-interactions-control-protein-expression-in-prokaryotes-5-2048.jpg)
![Do ubiquitous and abundant RNAs influence translation?
Given that ncRNAs are among the most abundant RNAs in the cell
([ncRNA] >> [mRNA])
AND that RNAs frequently hybridise
THEN maybe stochastic interactions with mRNAs inhibit translation
Corley & Laederach (2016) Bioinformatics: Selecting against accidental RNA interactions. eLife.](https://image.slidesharecdn.com/talk-161115083940/75/Random-RNA-interactions-control-protein-expression-in-prokaryotes-14-2048.jpg)
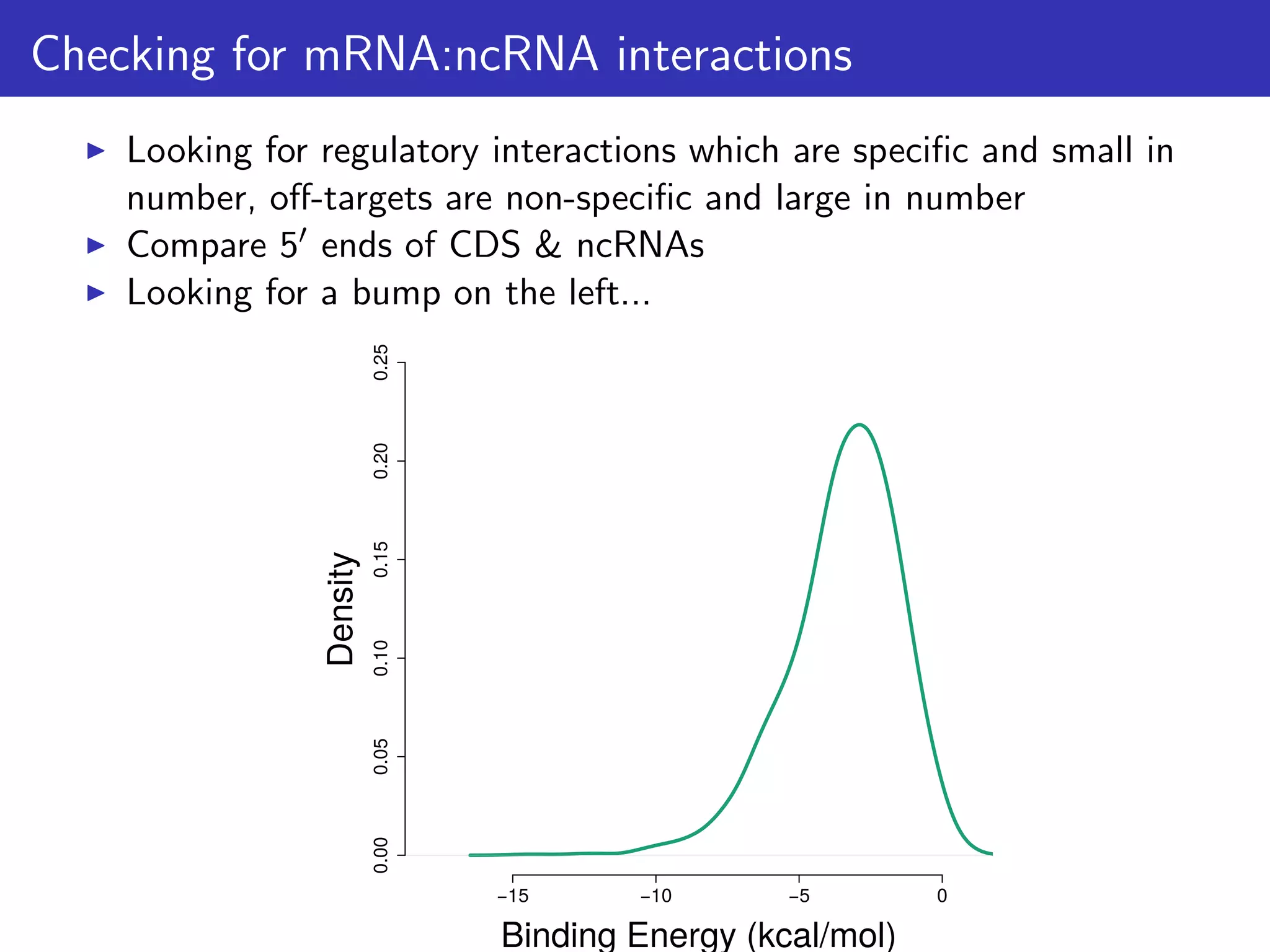
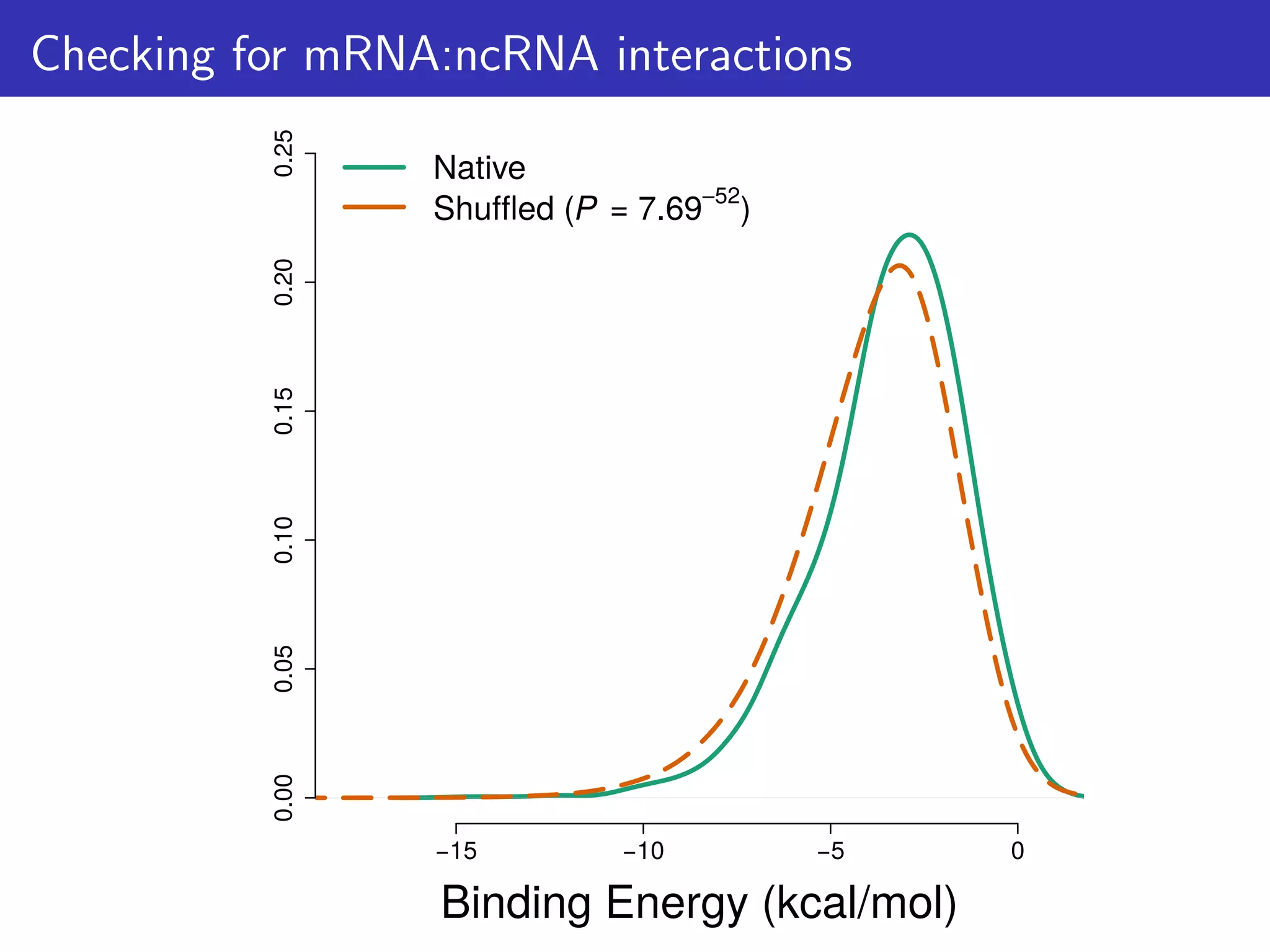
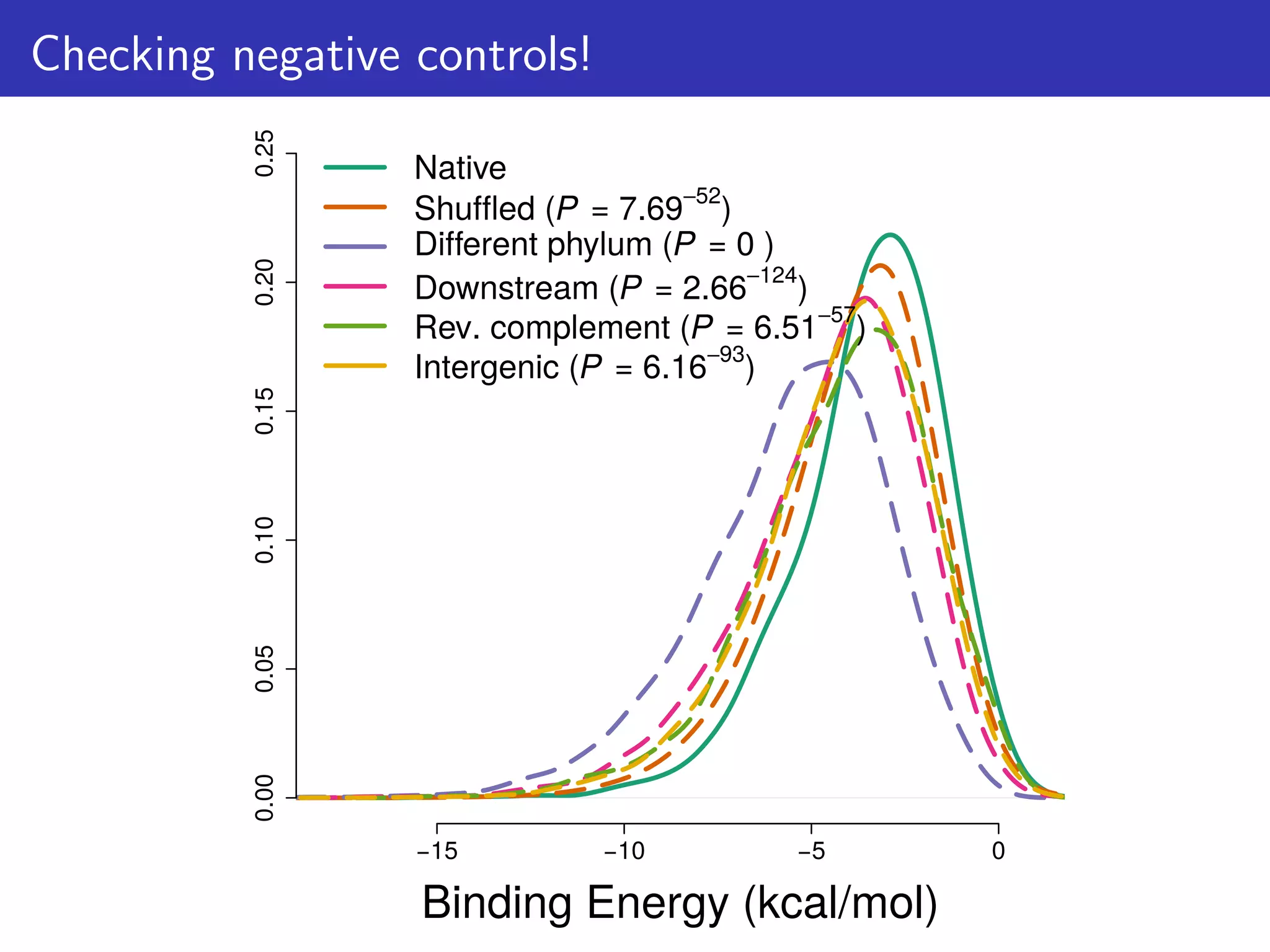
![How can this hypothesis be tested?
We predict that:
1. There is selection against mRNA:ncRNA interactions
2. That stochastic mRNA:ncRNA interactions influence [protein]:[mRNA]
ratios
For consistency: focus on 6 ncRNA families & 114 mRNAs/proteins
that are highly conserved & expressed; And first 21 nts of CDS.
Tested 1,582 bacterial & 118 archaeal genomes](https://image.slidesharecdn.com/talk-161115083940/75/Random-RNA-interactions-control-protein-expression-in-prokaryotes-15-2048.jpg)
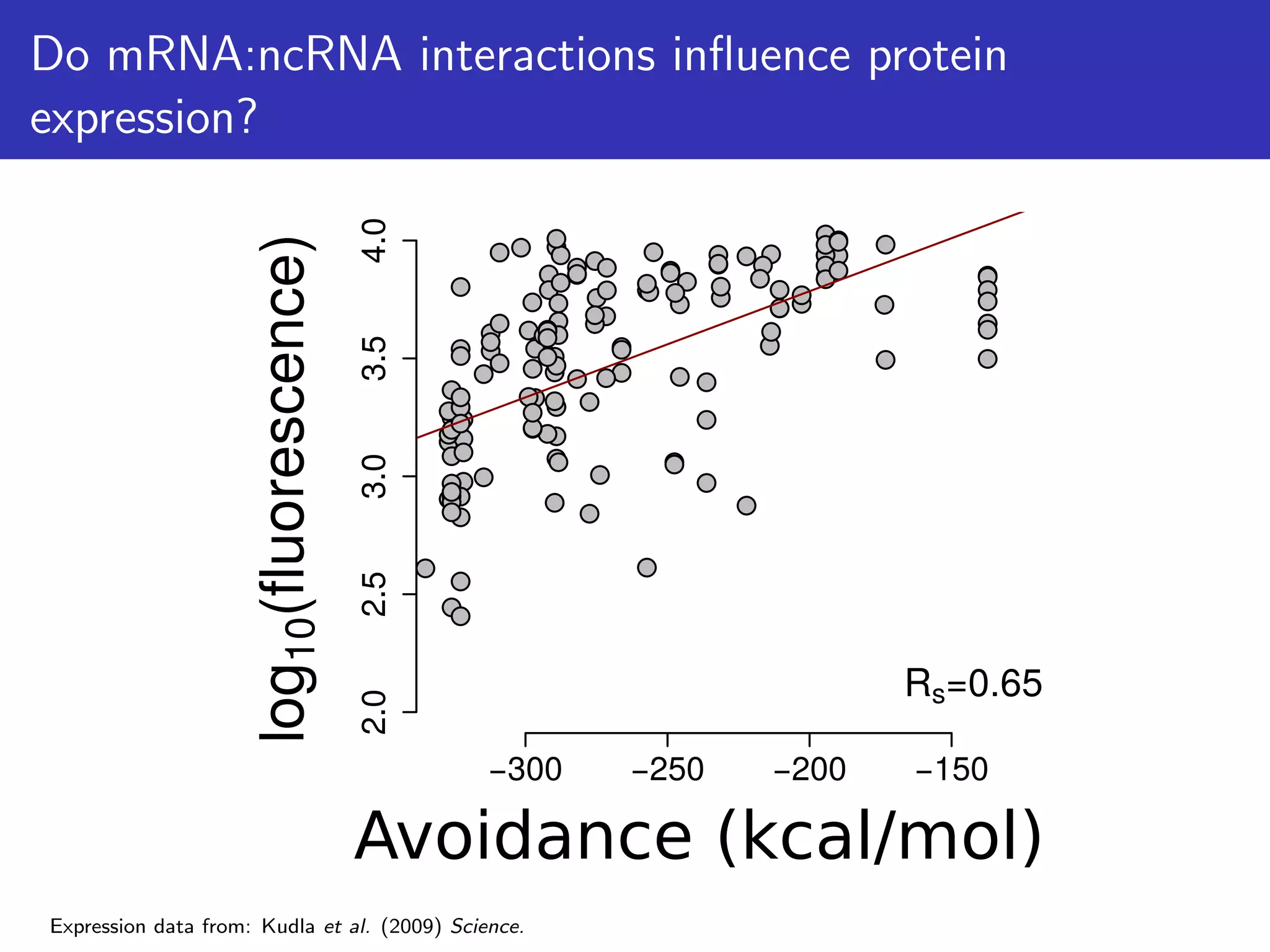
![Do mRNA:ncRNA interactions influence protein
expression?
Testing the relationship between protein abundance estimates and
avoidance, mRNA secondary structure, codon usage and mRNA
abundance
GFP datasets Mass-Spec datasets
E.coli
(n=52)
GFP/qPCR
E.coli
(n=154)
GFP/Northern
E.coli
(n=14,234)
mCherry/RNAseq
E.coli
(n=389)
MS/microarray
E.coli
(n=3,301)
MS/microarray
P.aeruginosa
(n=5,479)
MS/microarray
P.aeruginosa
(n=1,148)
MS/microarray
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*P < 0.05
0.0 0.60.2 0.4-0.2
Correlation Coefficient
Avoidance
Secondary
Structure
Codon
[mRNA]](https://image.slidesharecdn.com/talk-161115083940/75/Random-RNA-interactions-control-protein-expression-in-prokaryotes-18-2048.jpg)
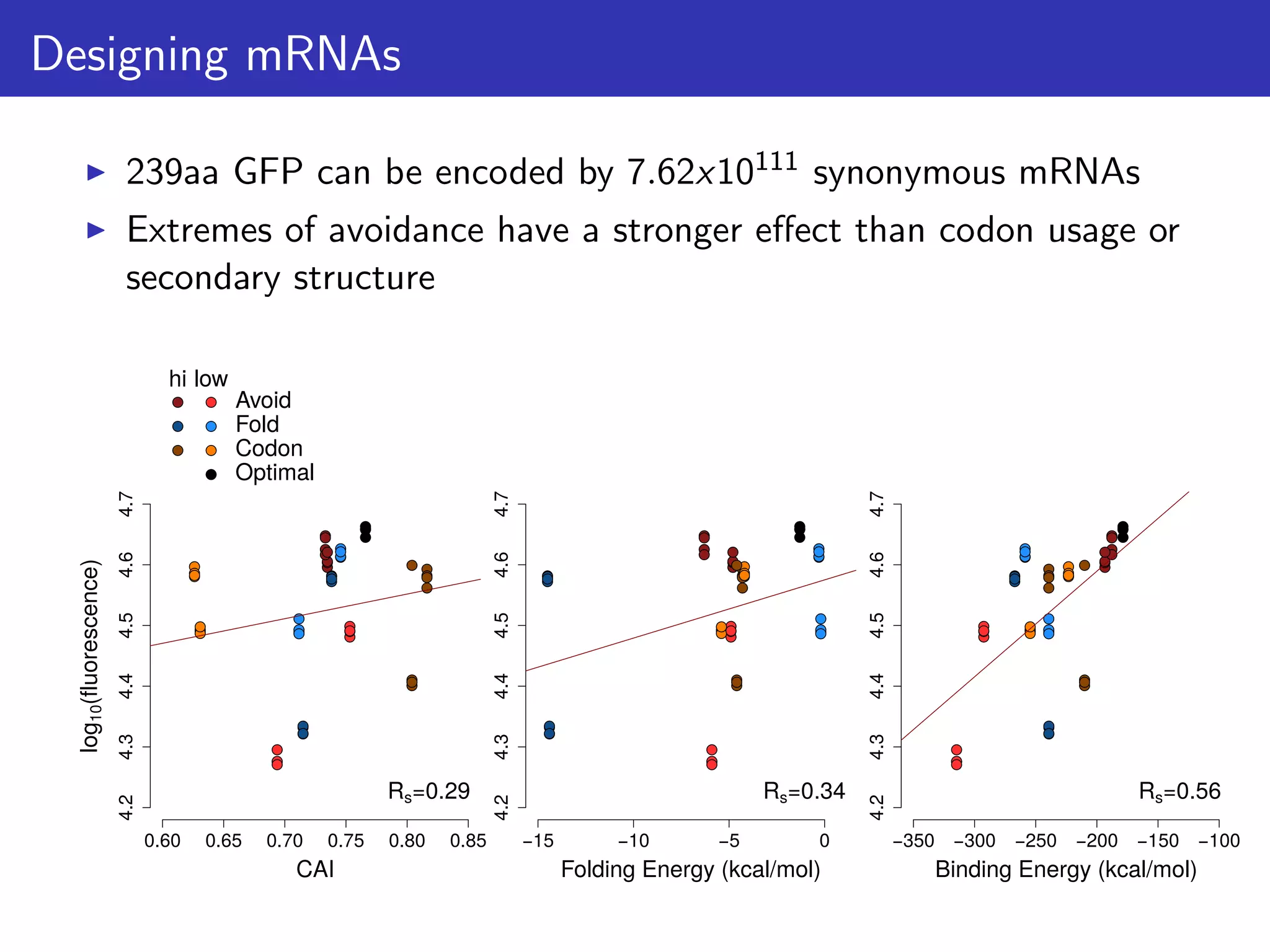
![Testing the extremes of expression
0.1
0.5
0.8
1.2
1.6
1.9
2.3
2.6
3
3.3
3.7
4.1
4.4
4.8
Freq
0
20
40
60
80
100
120
A
log10([Protein]/[mRNA])
Frequency
low expression (n=10)
high expression (n=10)
B
Avoidance
Codon
Sec.Str.
Null
Sec.Str.
Codon
Avoidance
−2
−1
0
1
2
*
*
Zscore
low expression (n=10)
high expression (n=10)
E. coli genes (n = 389)](https://image.slidesharecdn.com/talk-161115083940/75/Random-RNA-interactions-control-protein-expression-in-prokaryotes-19-2048.jpg)