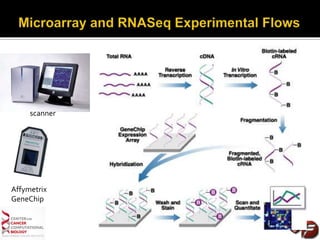
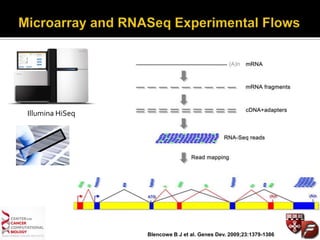
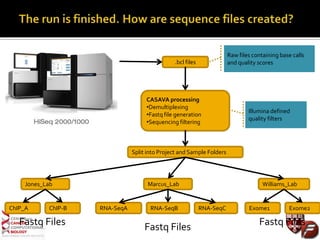
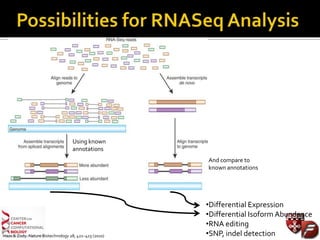
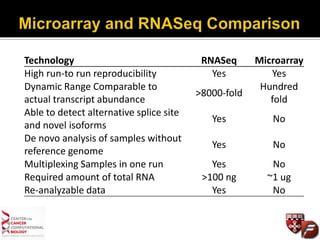
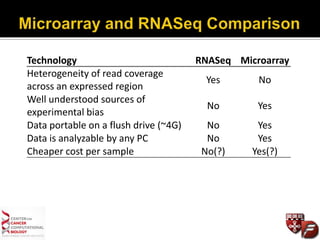
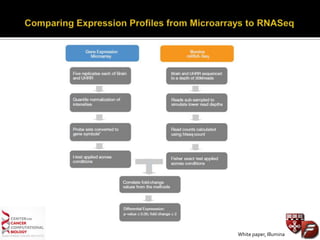
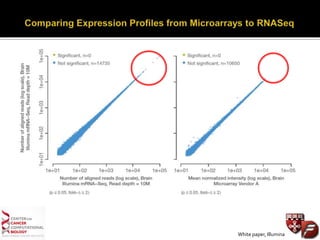
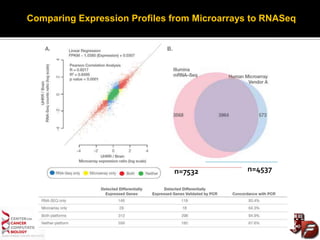
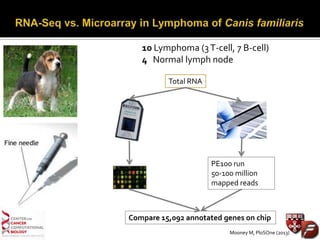
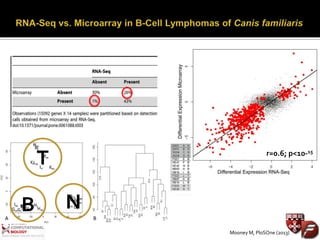
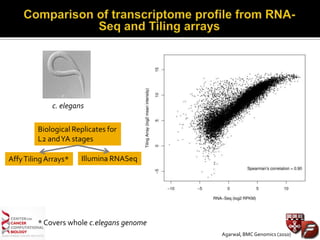
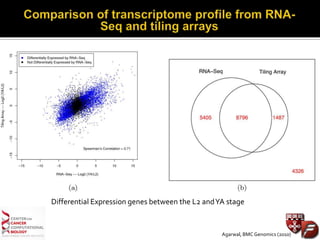
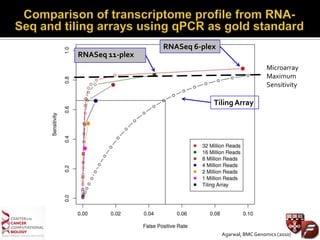
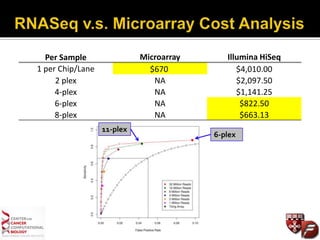
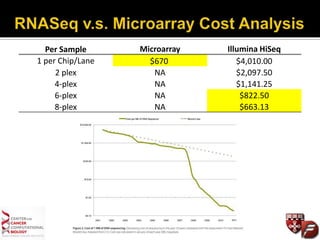
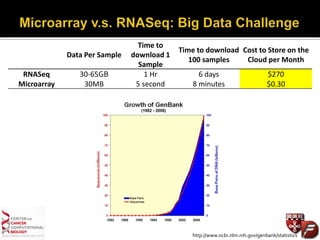
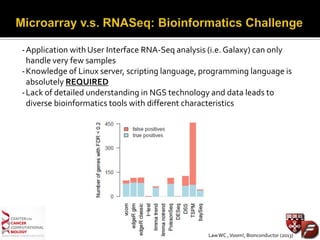
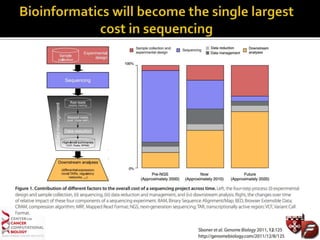
Transcriptome profiling using RNA-Seq or microarrays allows determination of differential gene expression between samples like normal vs. tumor. While RNA-Seq and microarrays are generally concordant, RNA-Seq provides more information like alternative splicing and novel transcripts but requires more computational resources. While the cost per sample of RNA-Seq is decreasing, storage and analysis of the large datasets requires specialized infrastructure.