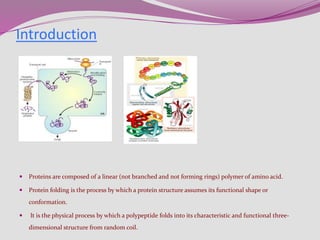
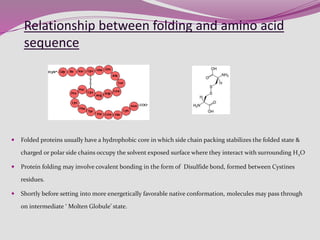
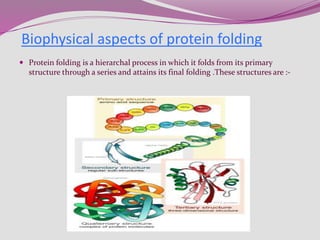
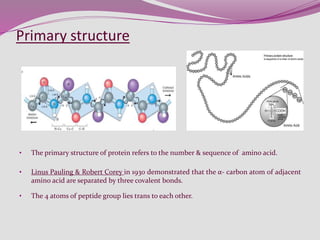
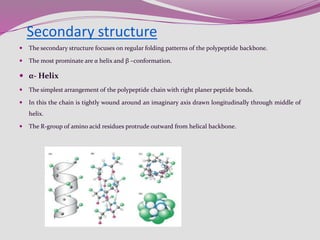
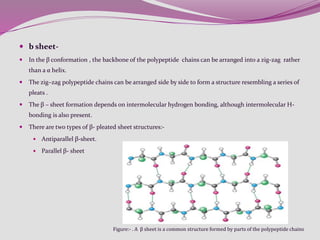
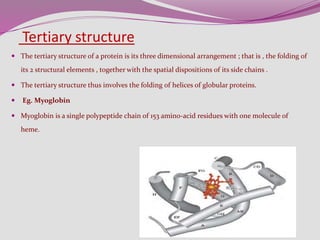
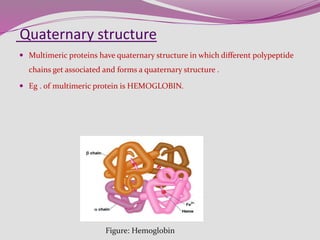
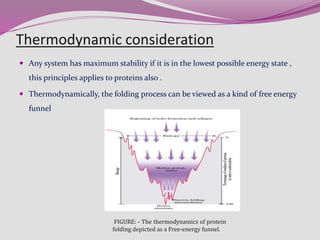
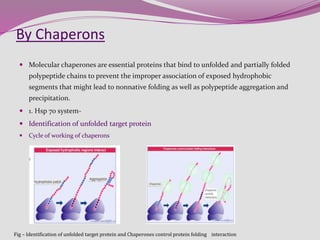
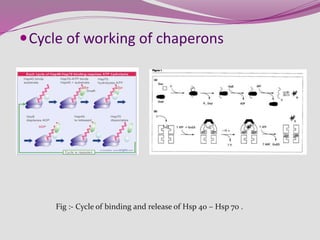
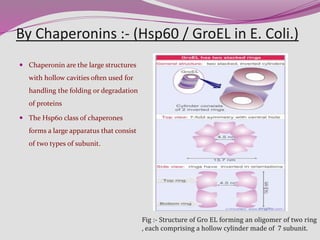
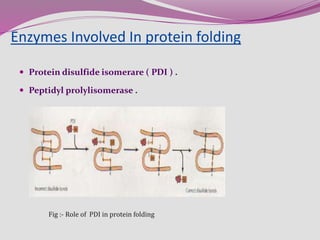
The document discusses protein folding, a process by which linear polypeptides achieve their functional three-dimensional structures from random coils. It addresses the Levinthal paradox, the hierarchical structure of proteins (primary, secondary, tertiary, quaternary), and the roles of chaperones and enzymes in facilitating proper folding. Additionally, it emphasizes the importance of correct folding for protein functionality and the potential consequences of misfolding.