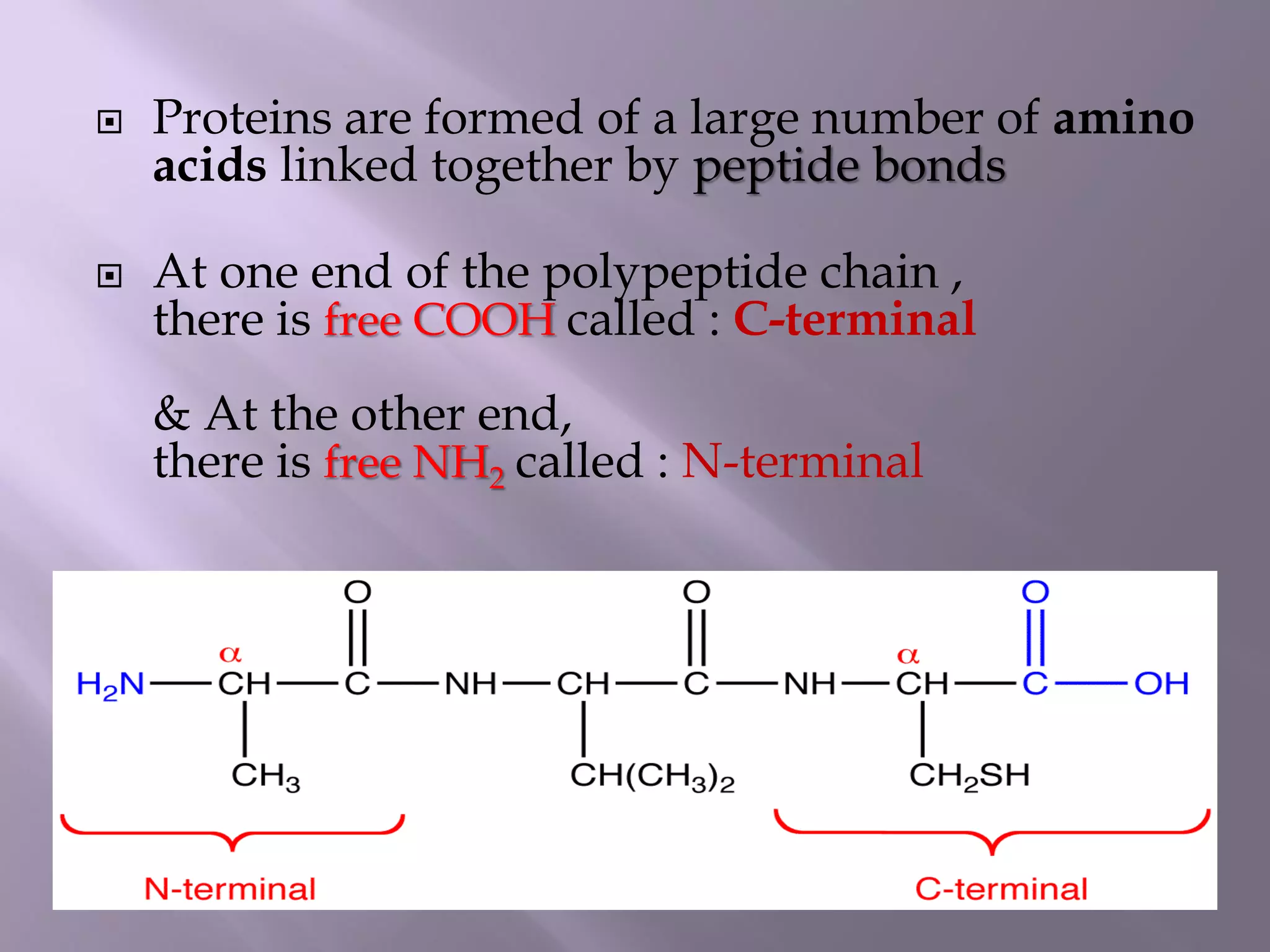
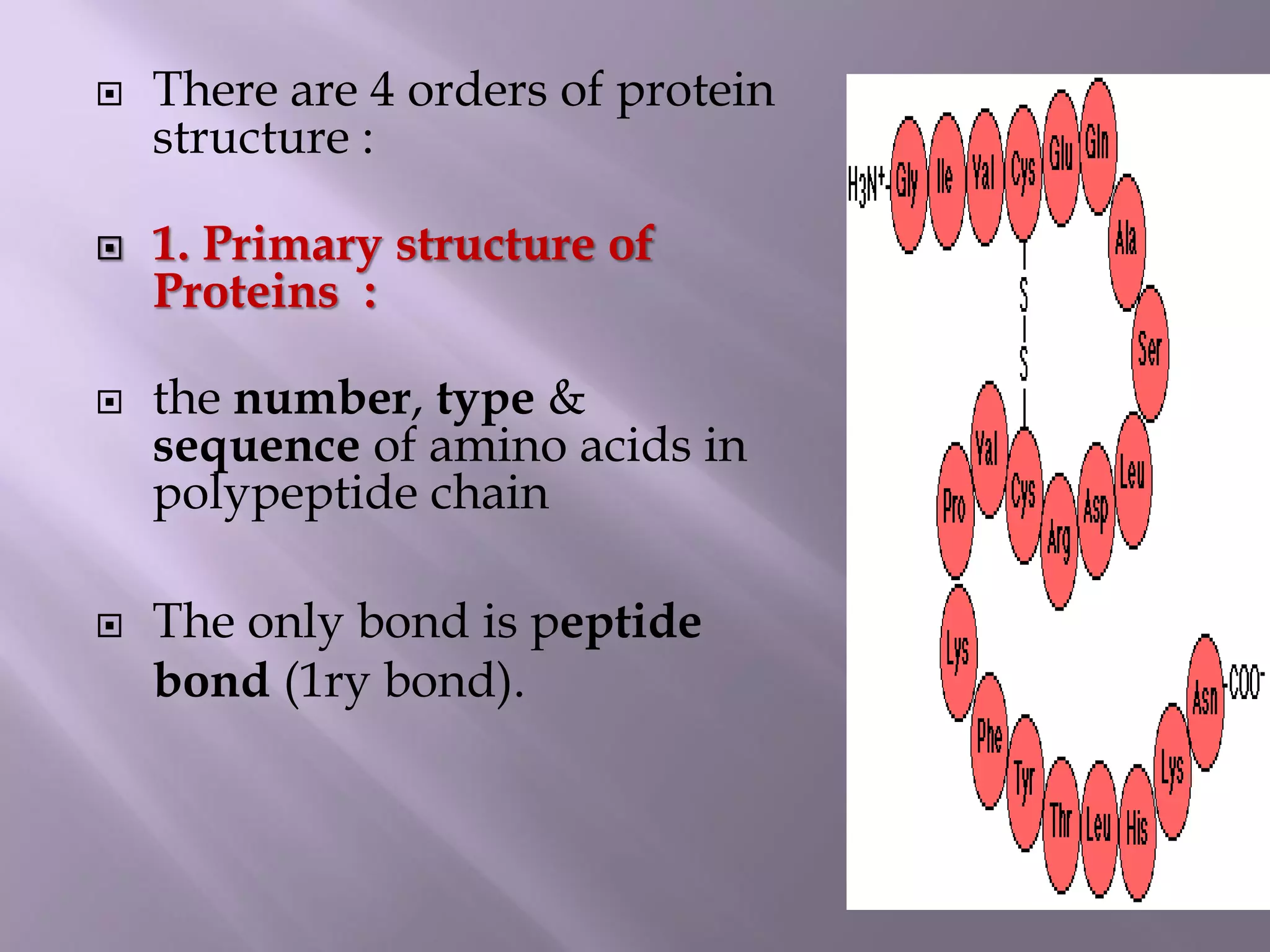
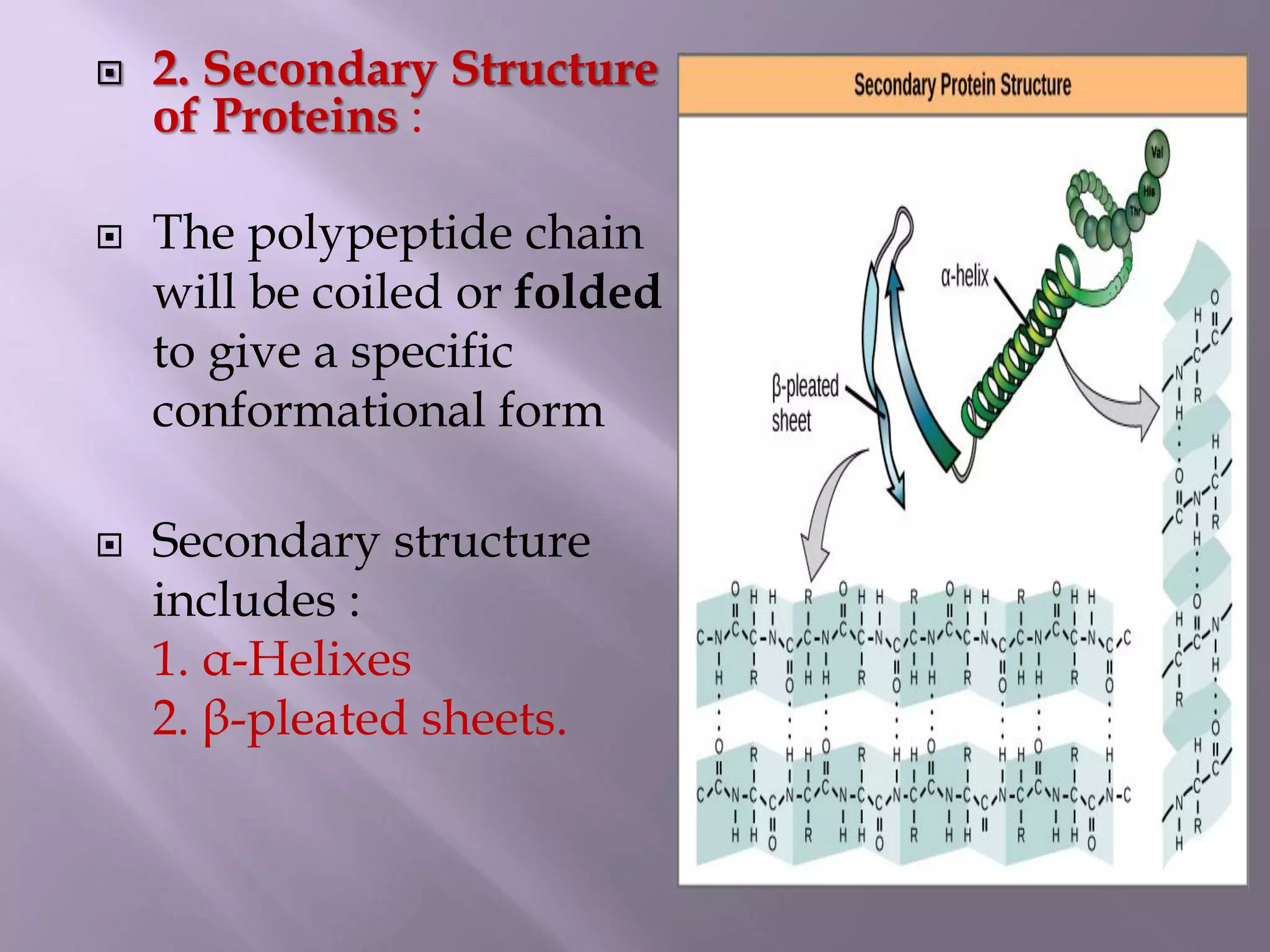
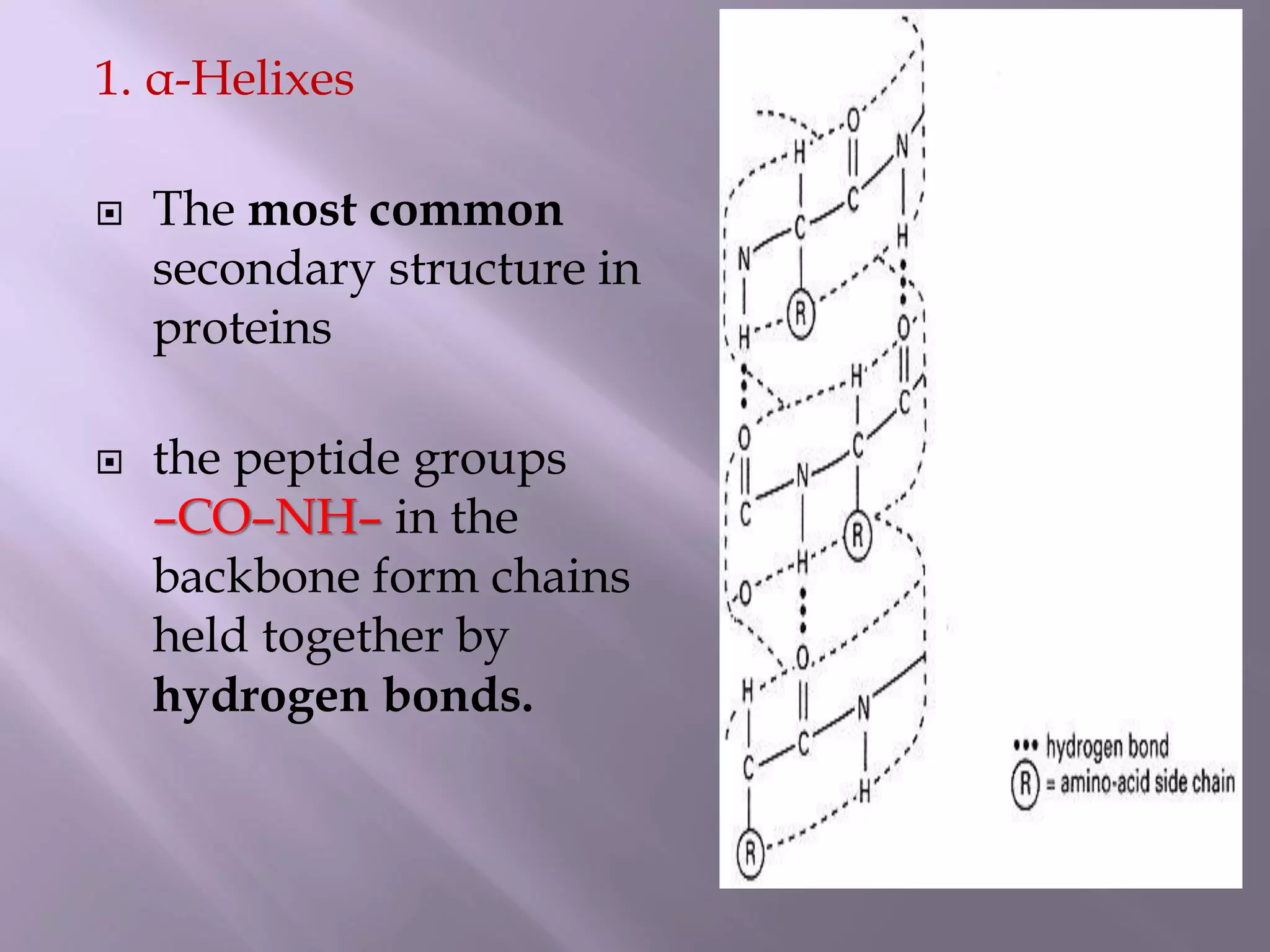
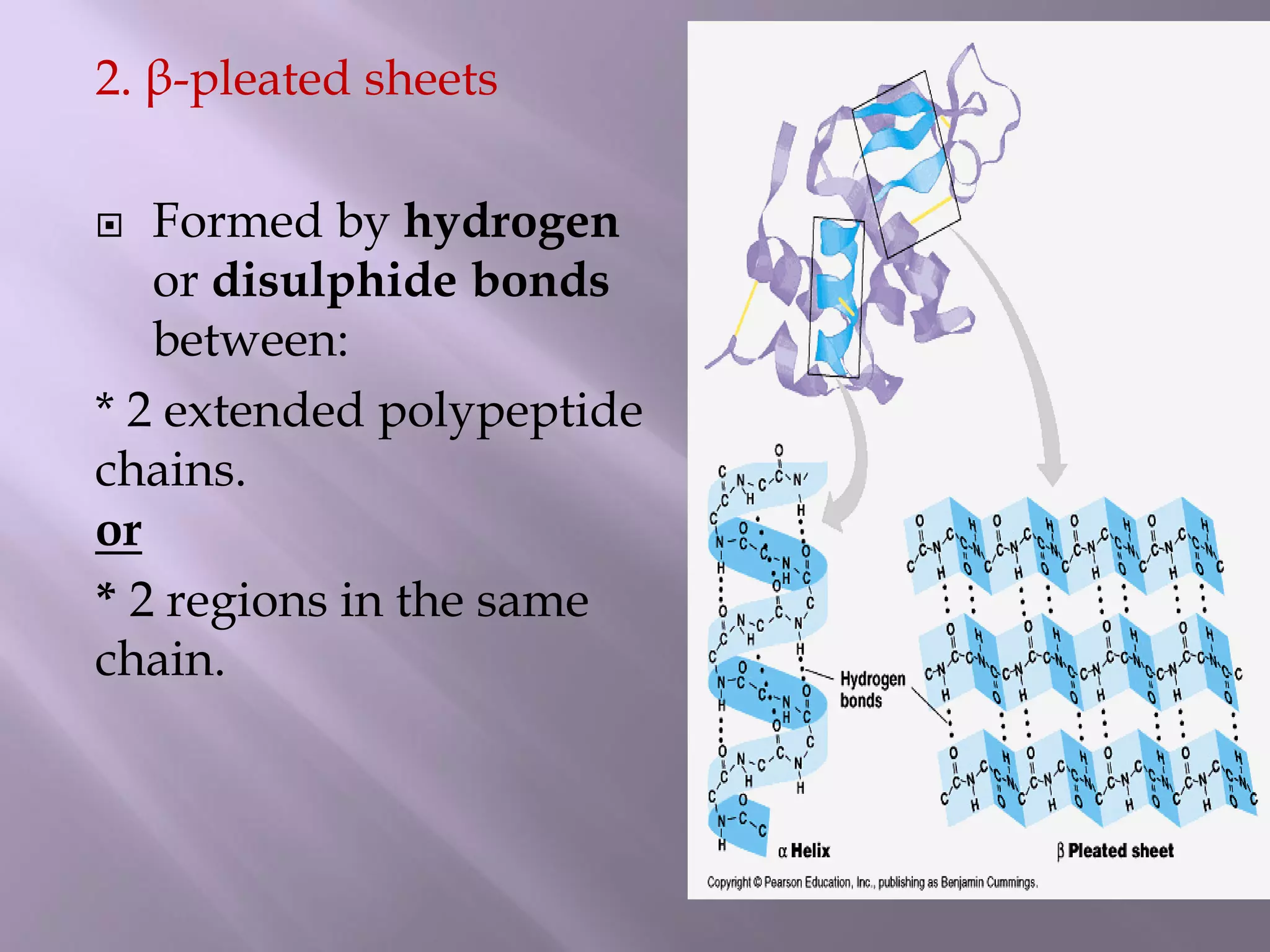
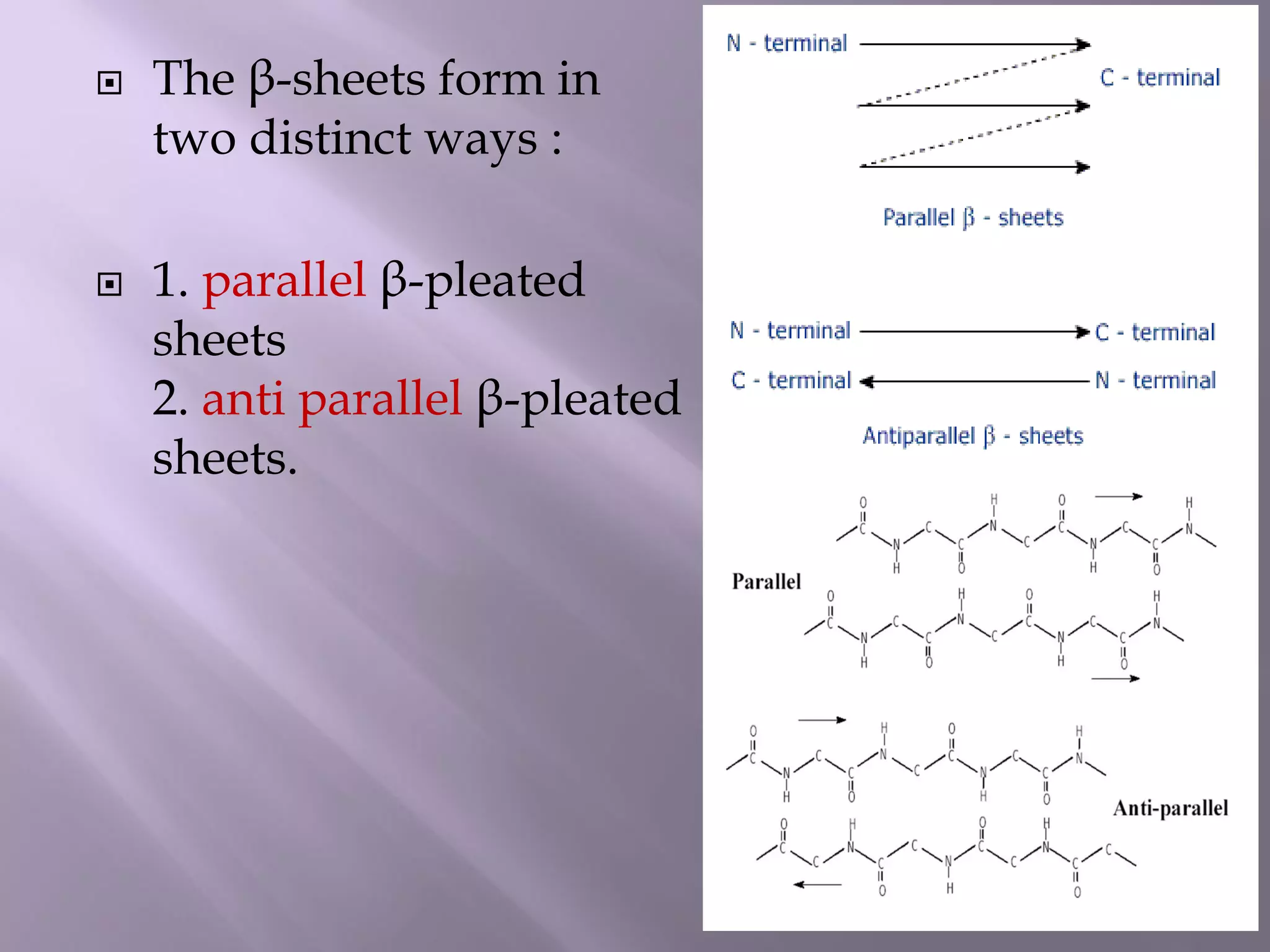
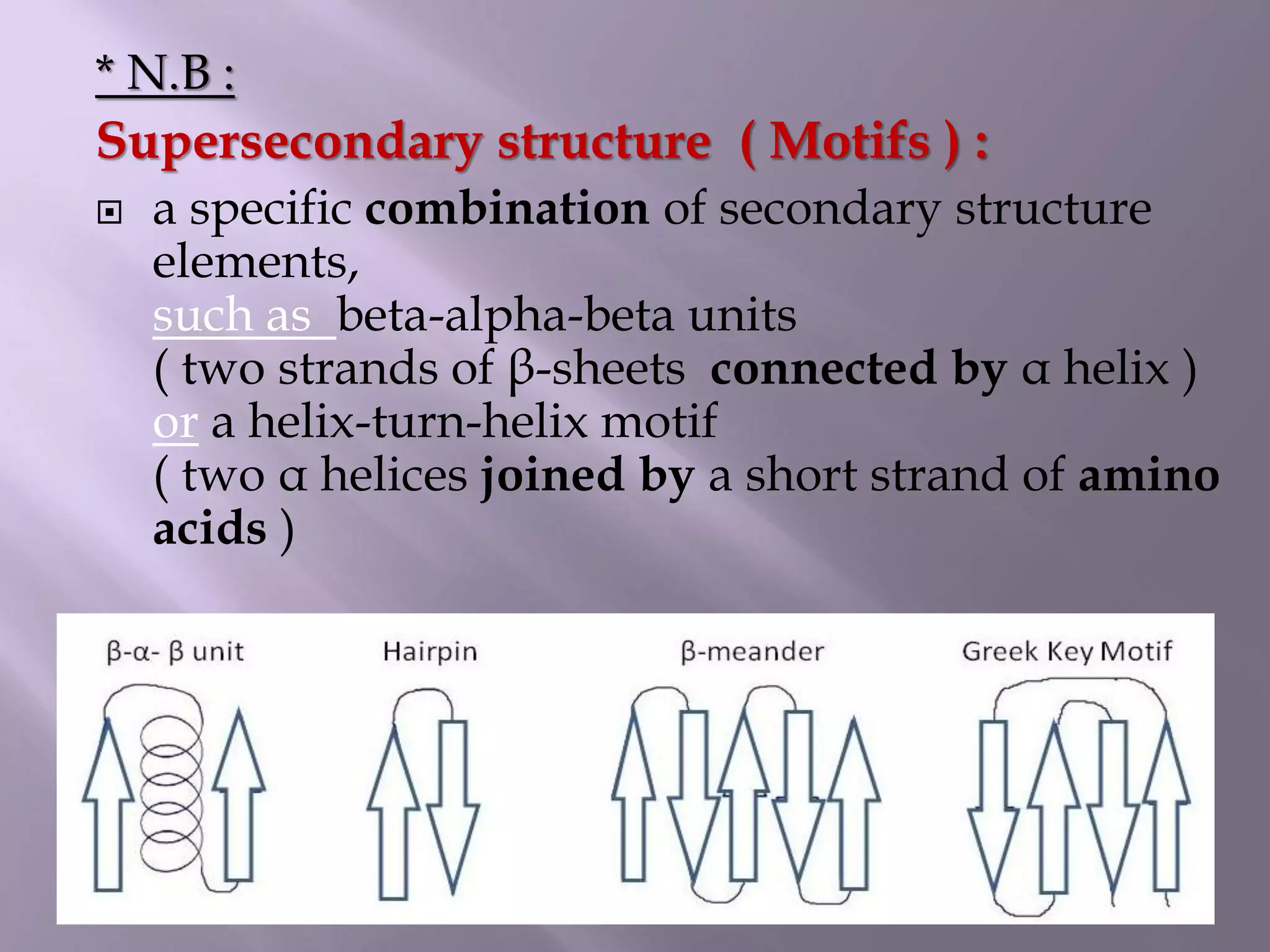
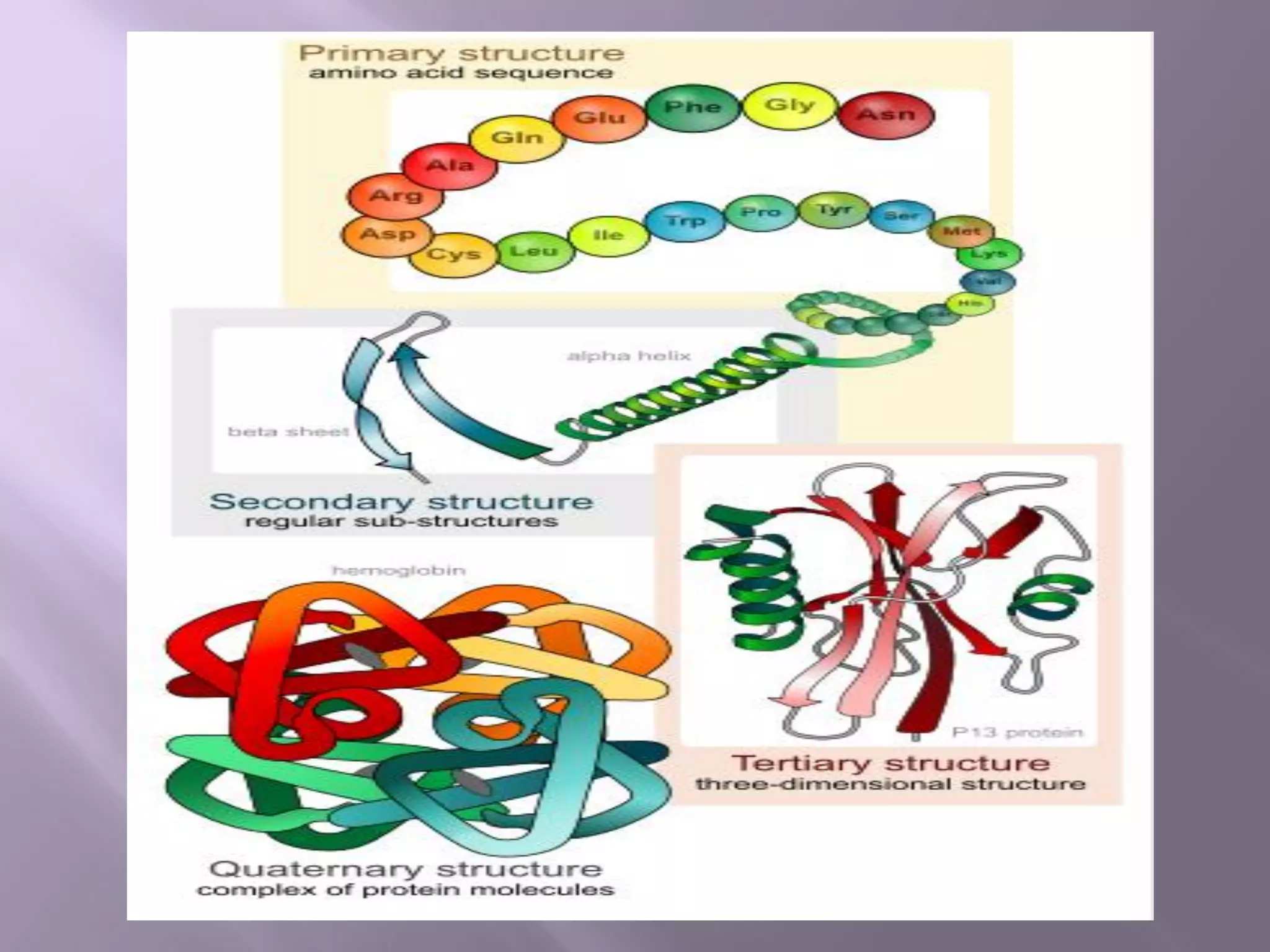
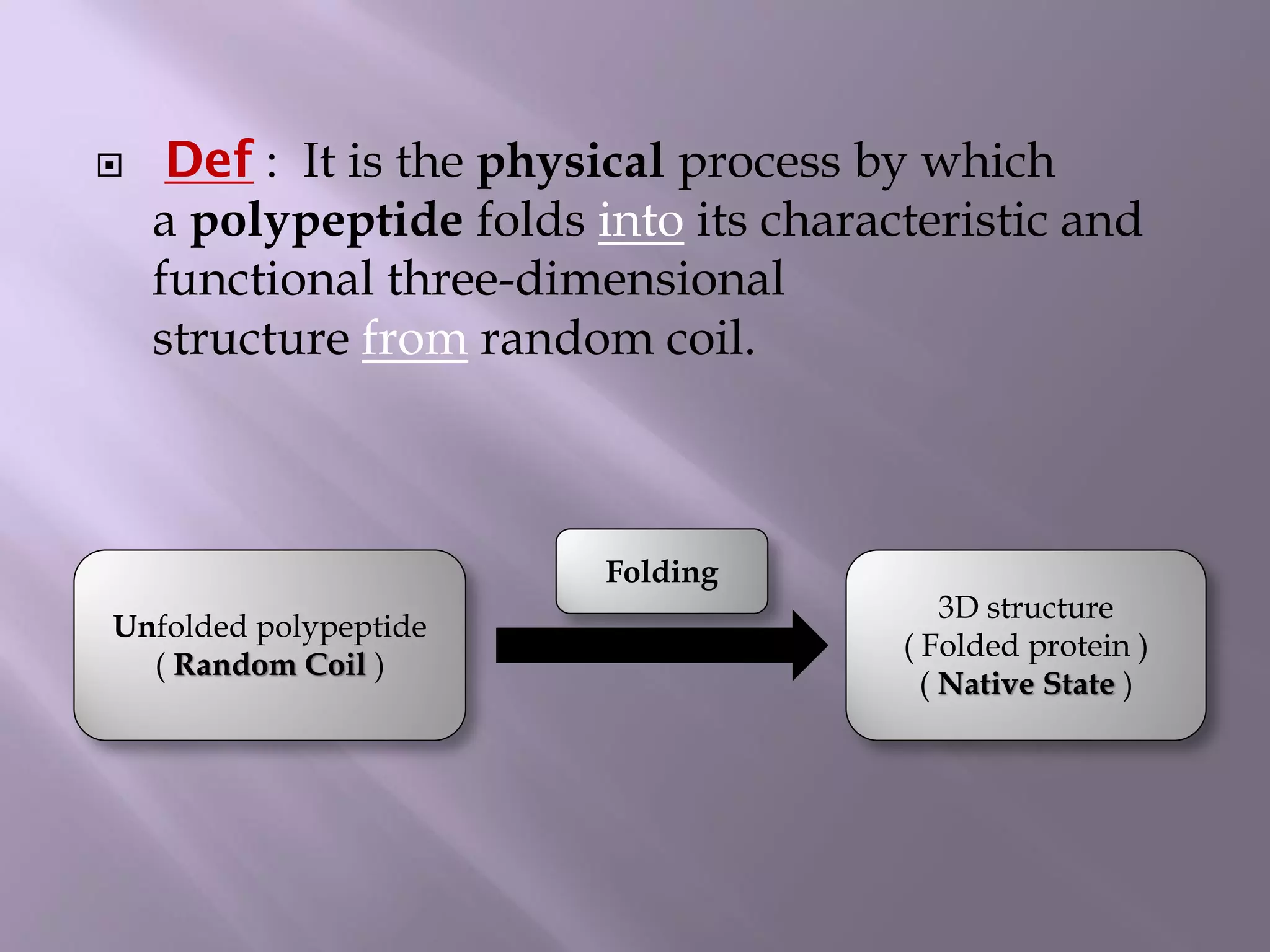
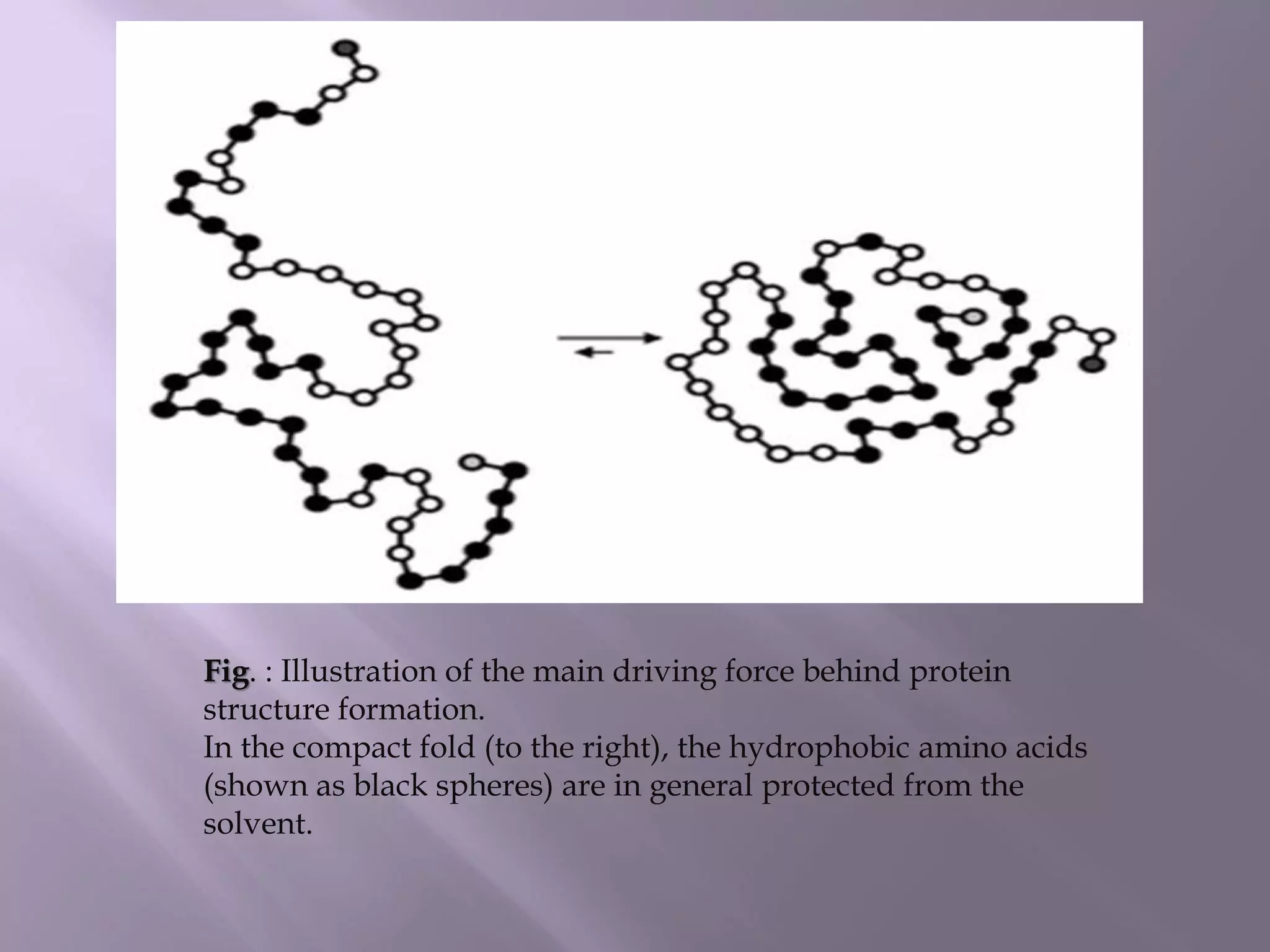
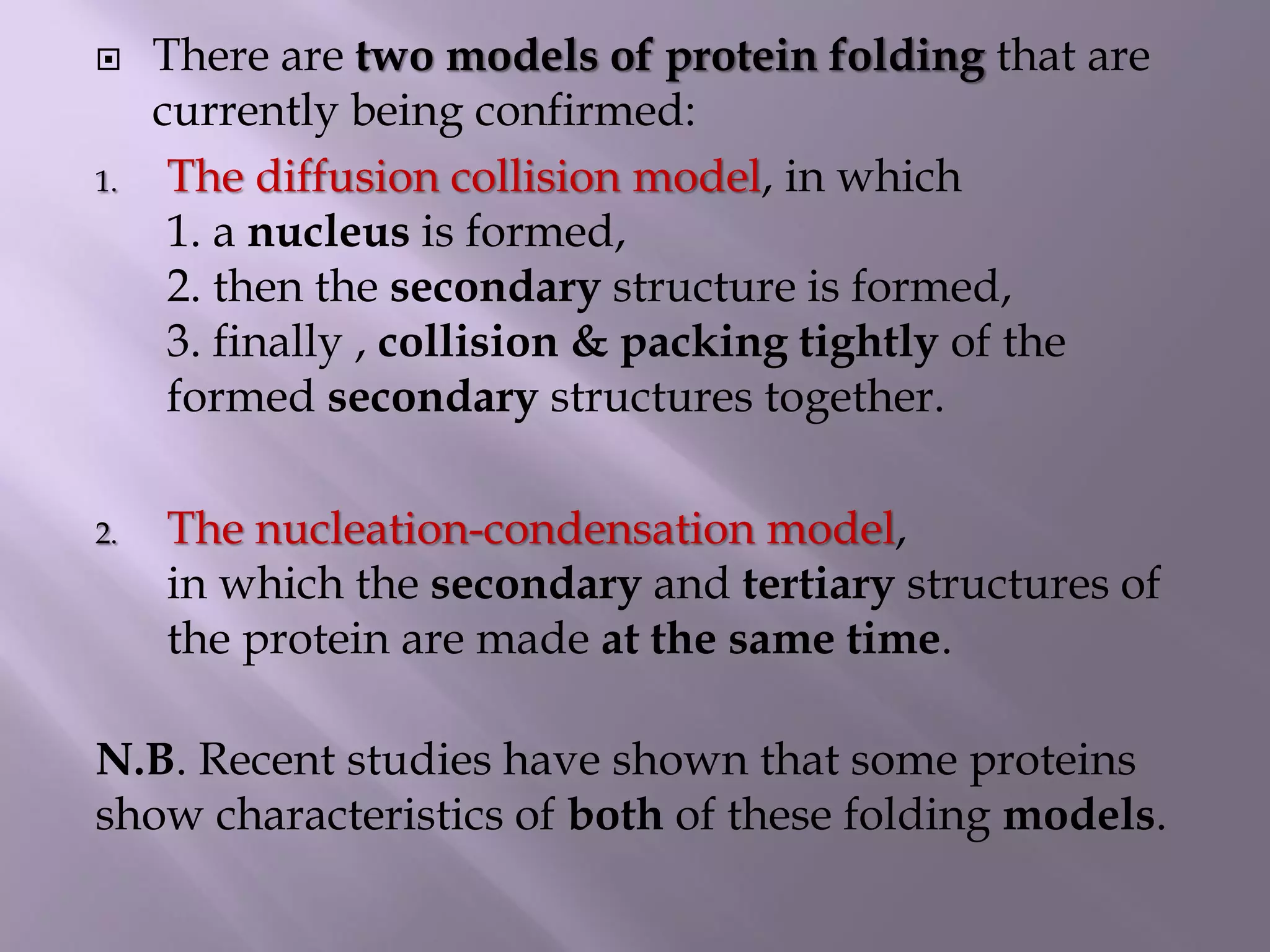
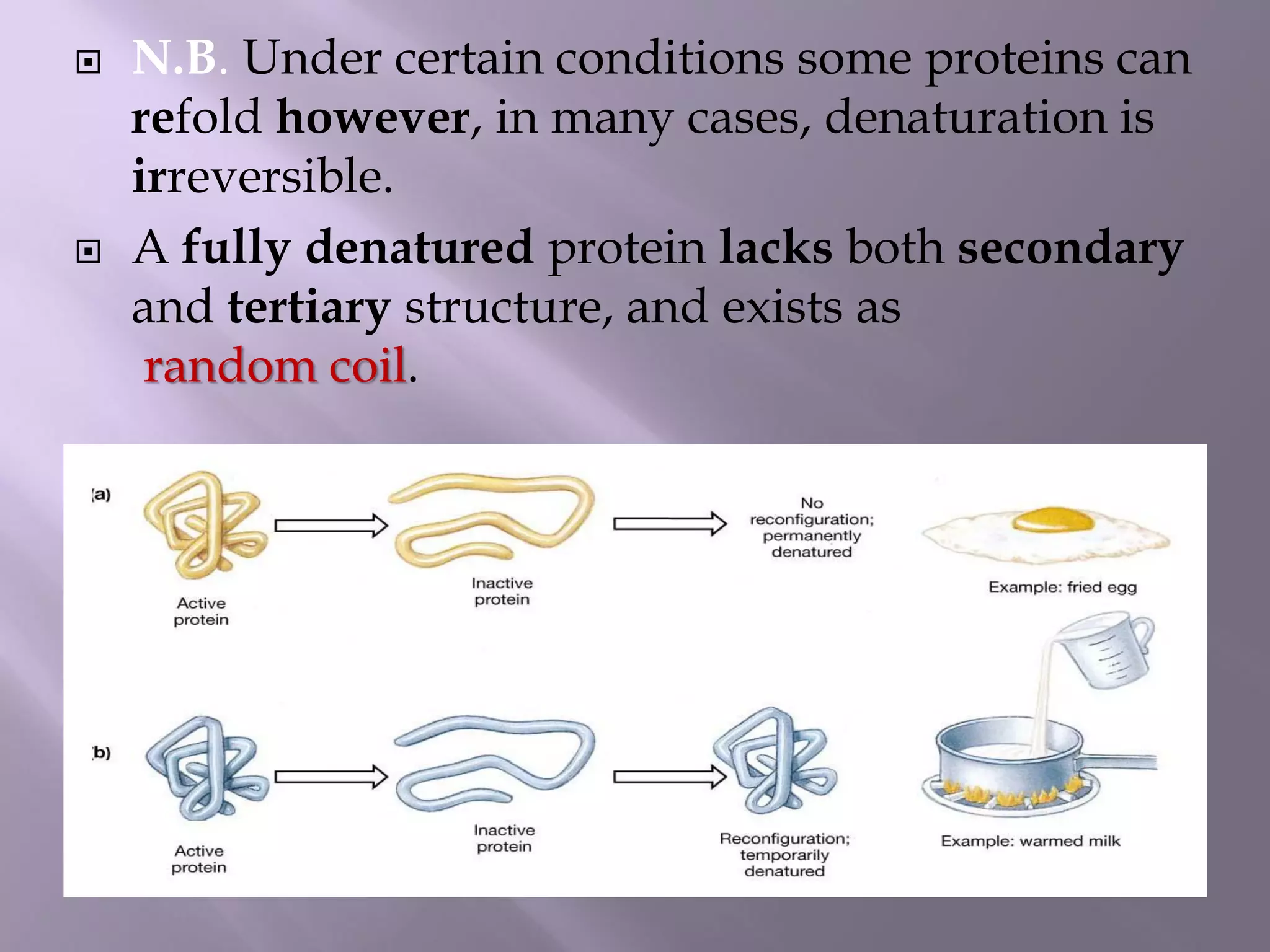
The document discusses protein folding, its structures (primary, secondary, tertiary, and quaternary), and the relationship between amino acid sequences and their functionality. It highlights the role of environmental factors and molecular chaperones in facilitating proper folding, as well as the consequences of misfolded proteins, which can lead to various diseases. Additionally, it outlines different models of protein folding and the significance of maintaining the native state for biological activity.