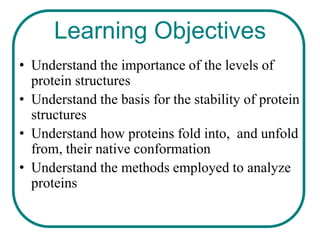
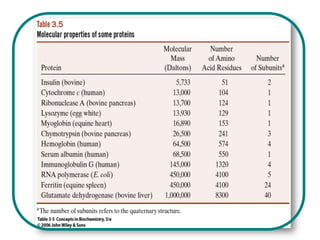
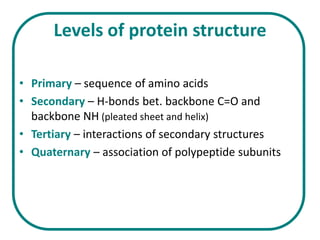
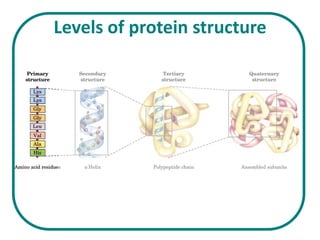
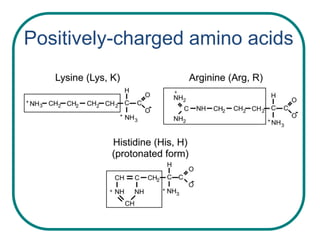
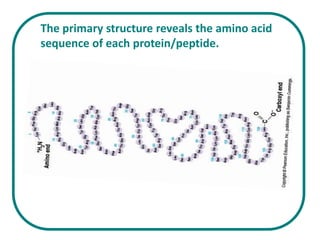
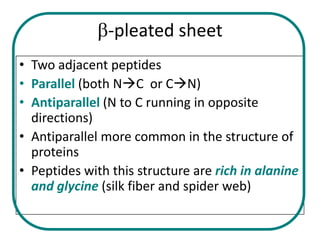
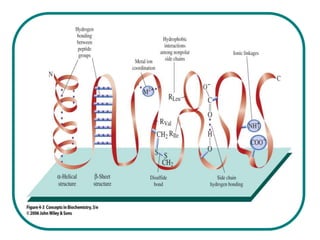
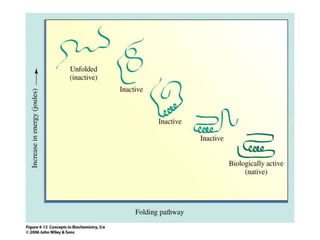
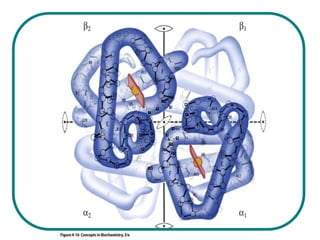
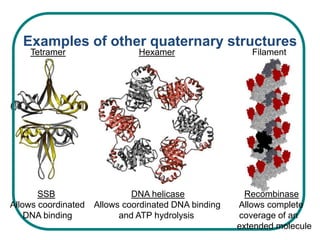
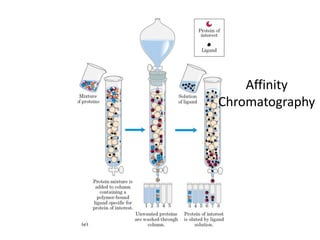
The document discusses the structure of proteins at multiple levels: primary, secondary, tertiary, and quaternary. The primary structure is the amino acid sequence. Secondary structures include alpha helices and beta sheets formed by hydrogen bonds. Tertiary structure involves the folding of secondary structures into compact 3D shapes stabilized by interactions like hydrophobic effects. Quaternary structure refers to the arrangement of multiple polypeptide subunits in a single protein. The document also covers protein classification, folding, isolation, and sequencing techniques.