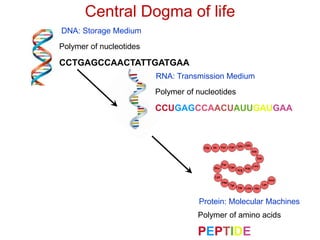
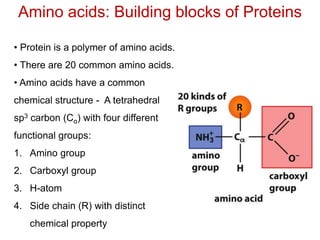
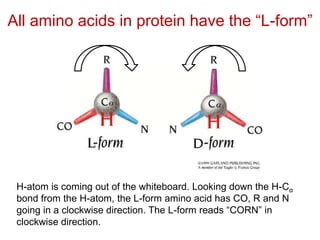
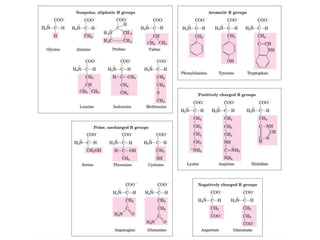
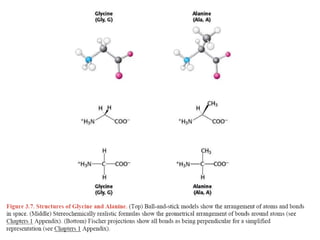
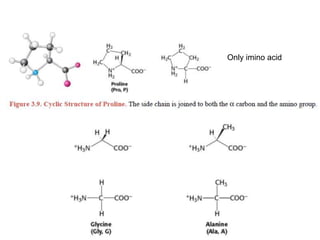
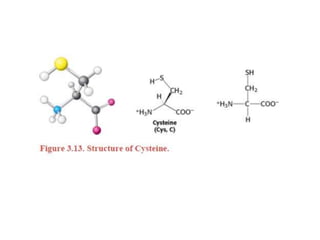
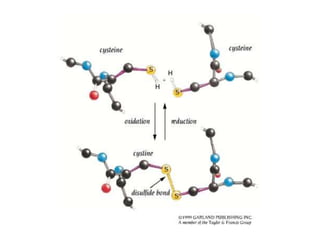
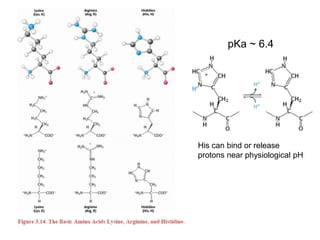
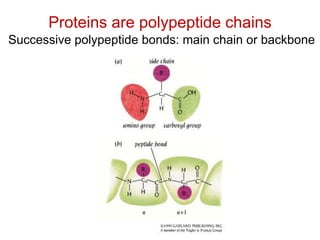
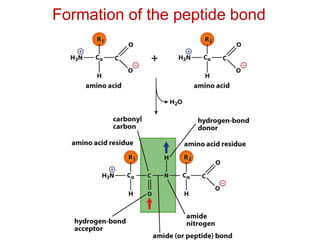
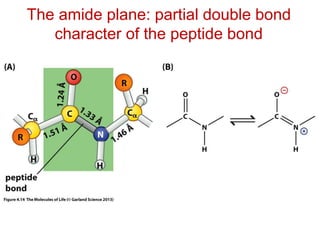
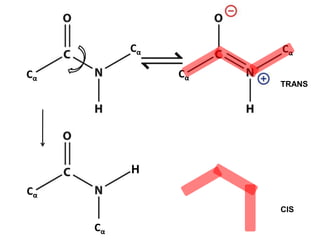
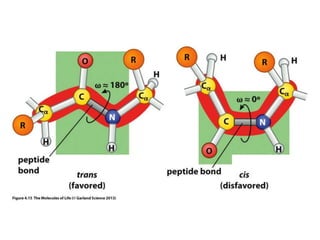
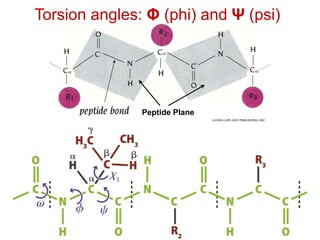
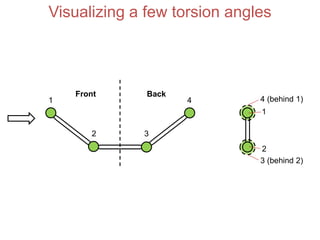
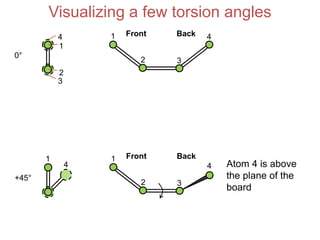
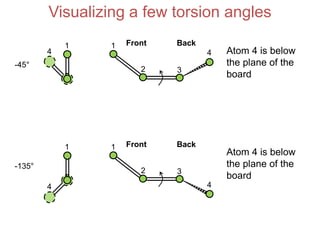
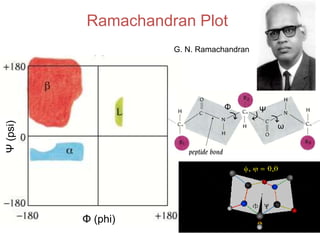
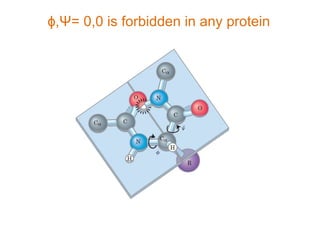
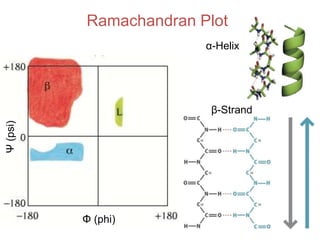
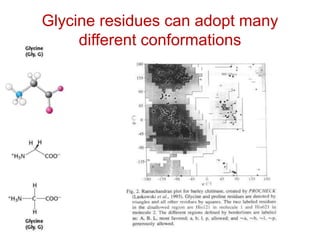
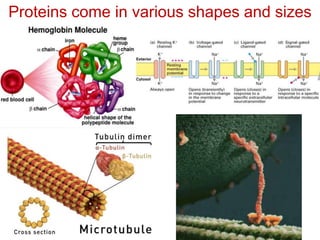
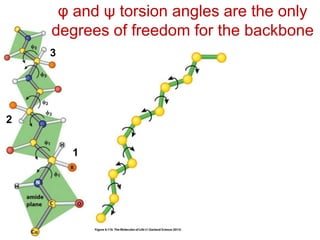
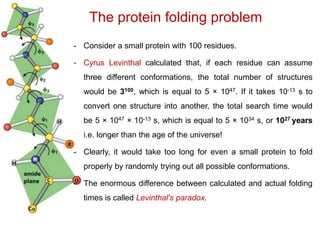
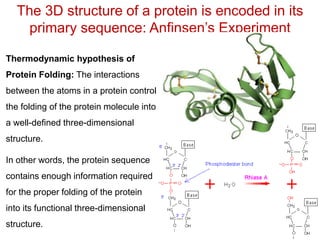
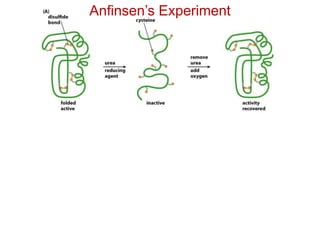
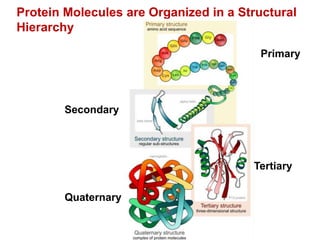
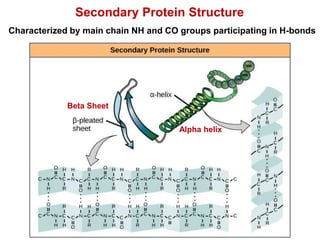
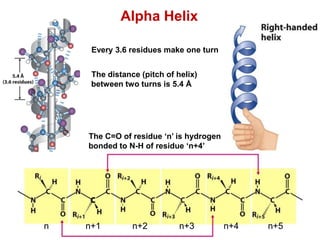
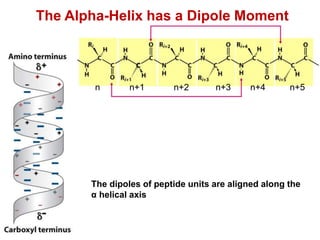
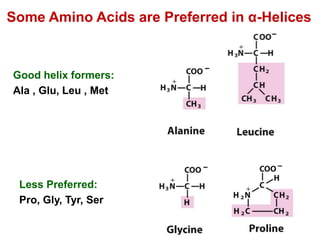
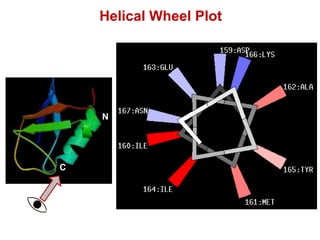
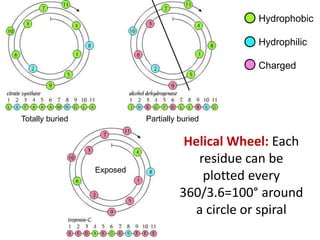
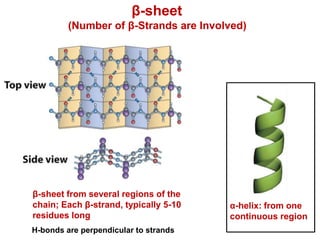
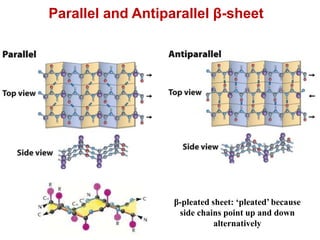
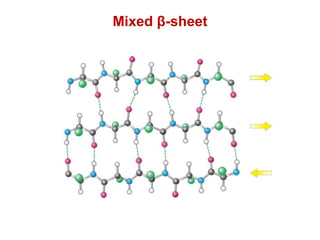
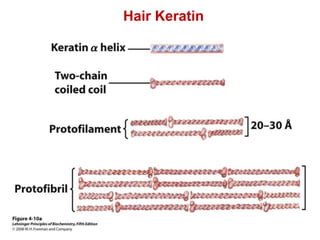
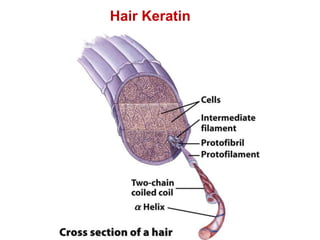
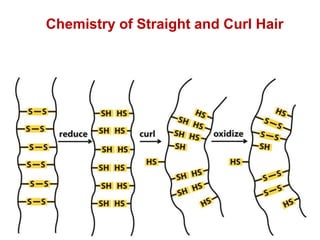
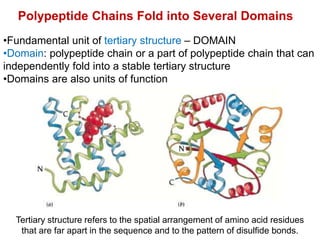
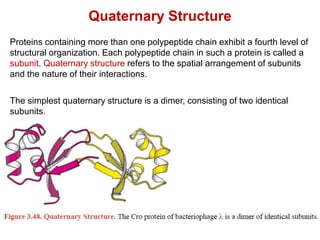
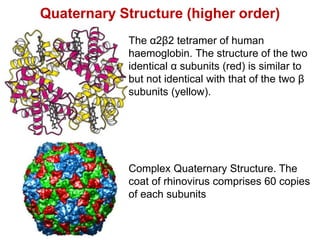
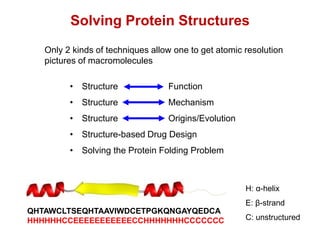
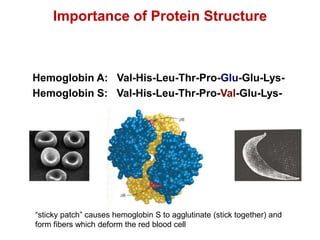
This document provides an overview of protein structure and function. It discusses the central dogma of life, the four levels of protein structure (primary, secondary, tertiary, and quaternary), common secondary structures like alpha helices and beta sheets, protein folding, domains, and important experiments like Anfinsen's that demonstrated proteins can fold on their own. It also mentions applications of protein structure knowledge like structure-based drug design and solving medical problems like sickle cell anemia.