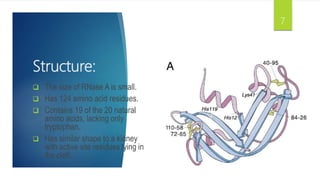
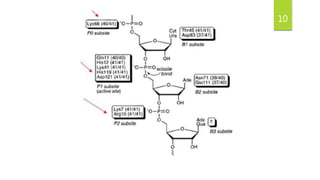
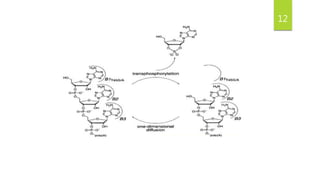
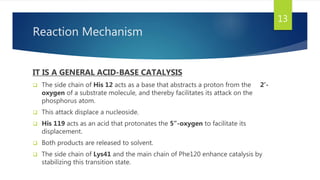
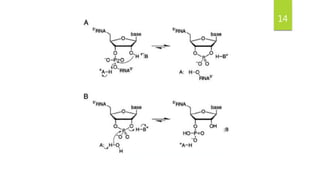
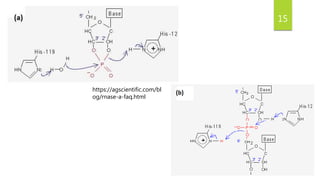
Ribonuclease A (RNase A) is an enzyme that catalyzes the degradation of RNA. It does so through acid-base catalysis using histidine residues to cleave phosphodiester bonds in RNA. RNase A has been extensively studied due to its small size and stability, yielding insights into protein structure and folding. It specifically binds and degrades RNA, with substrate binding involving interactions between RNA bases and enzymatic subsites in RNase A.