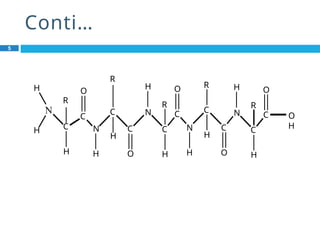
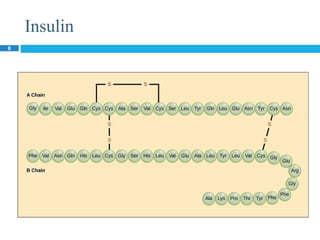
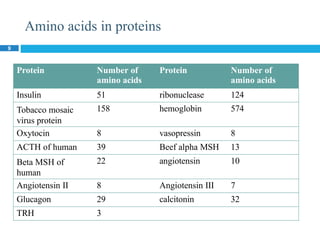
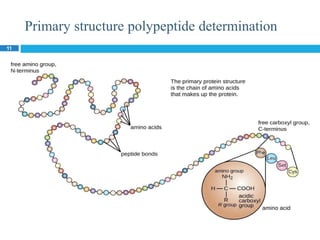
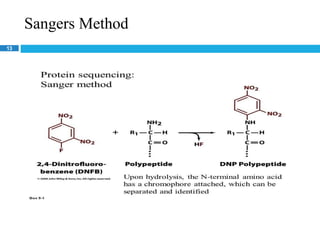
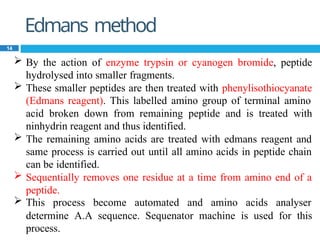
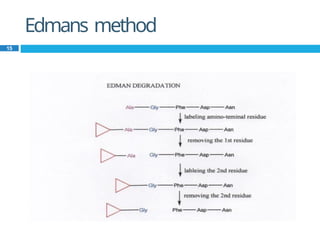
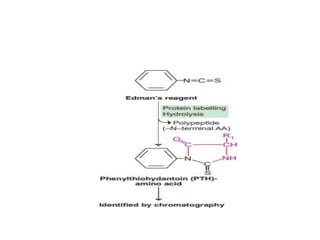
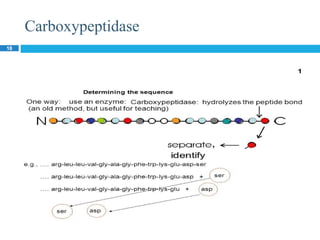
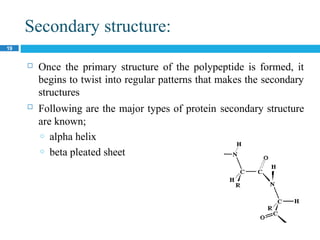
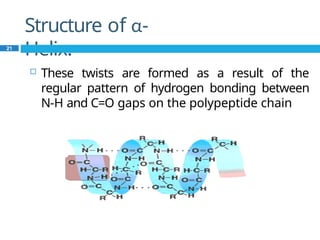
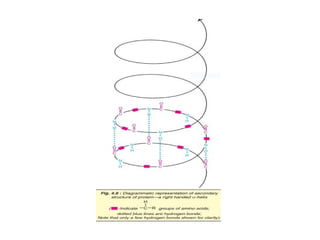
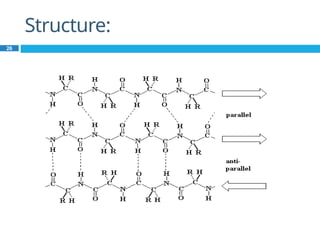
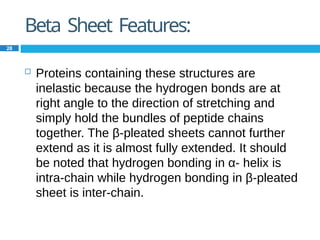
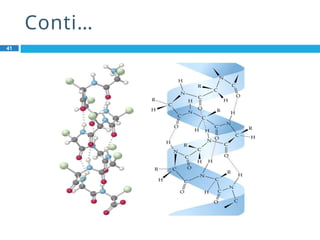
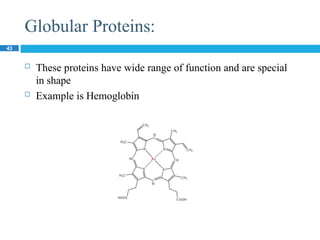
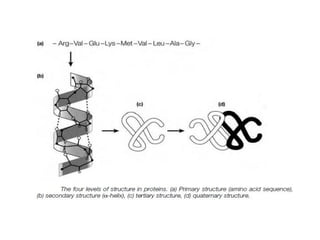
The document provides an in-depth overview of protein structure and function, detailing the organizational levels of proteins including primary, secondary, tertiary, and quaternary structures. It emphasizes the role of amino acids, the importance of specific sequences for proper function, and methods of determining polypeptide sequences. Additionally, it outlines various roles of proteins in biological processes, such as enzymes, oxygen transport, and tissue structure.