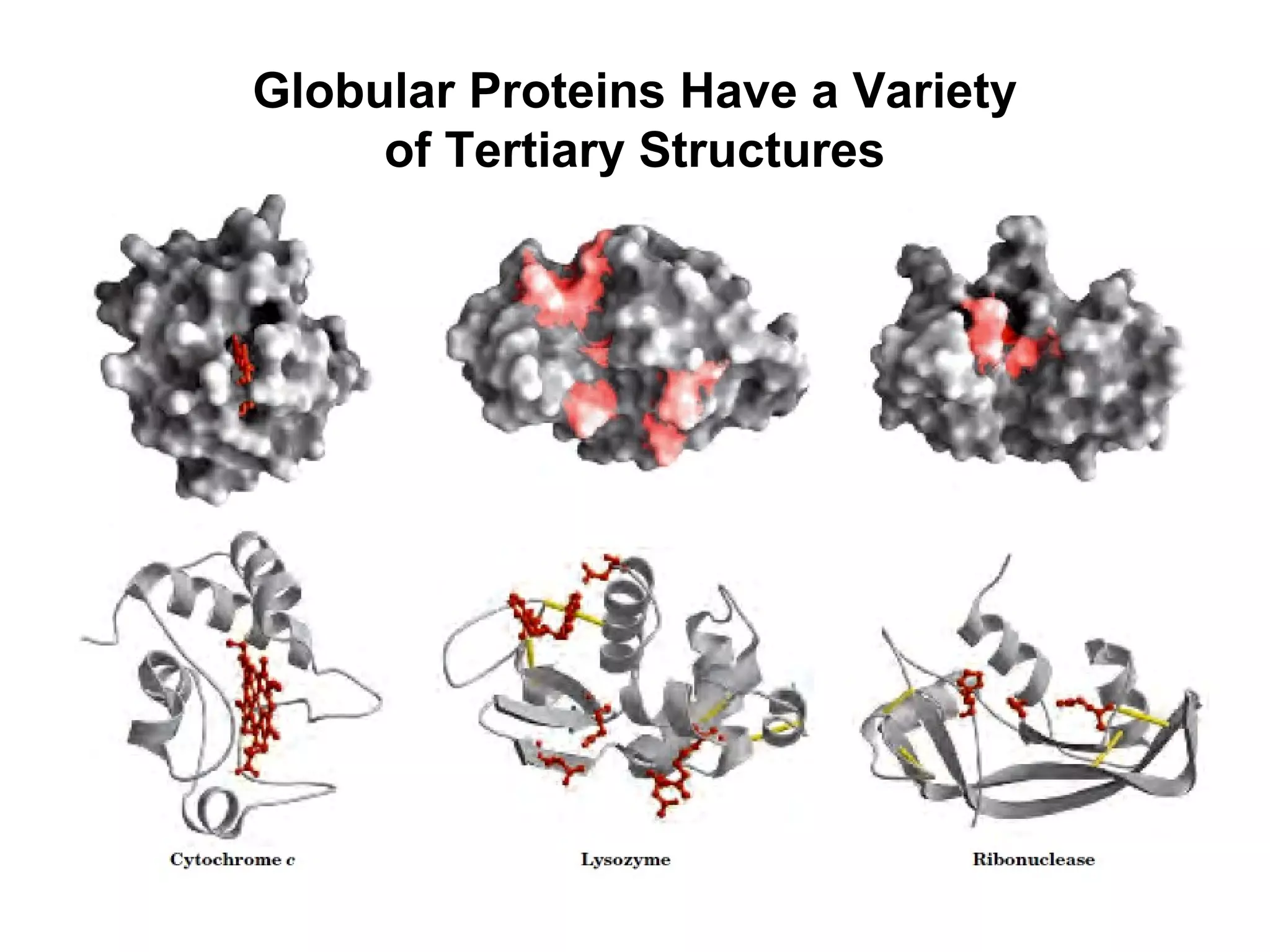
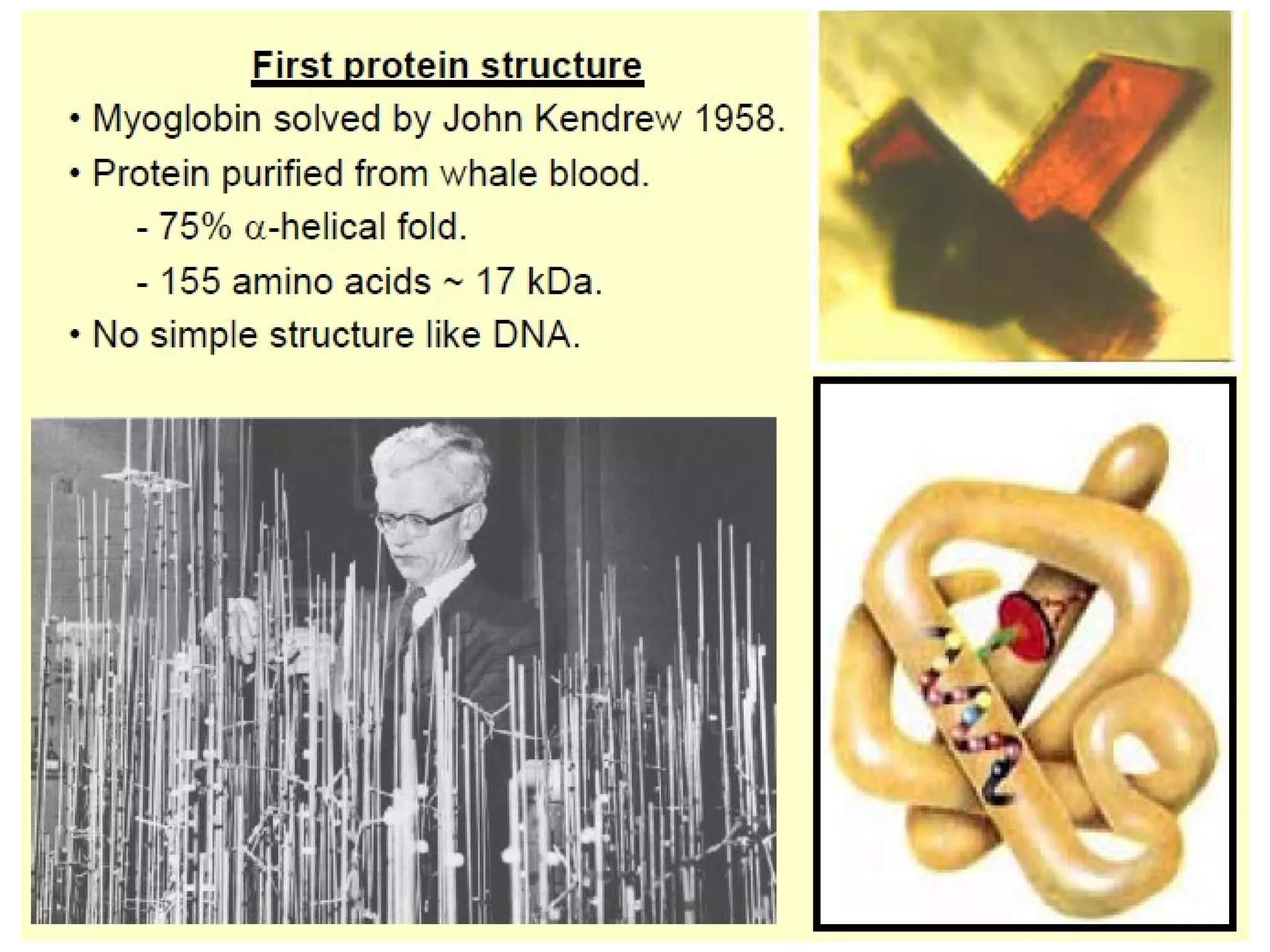
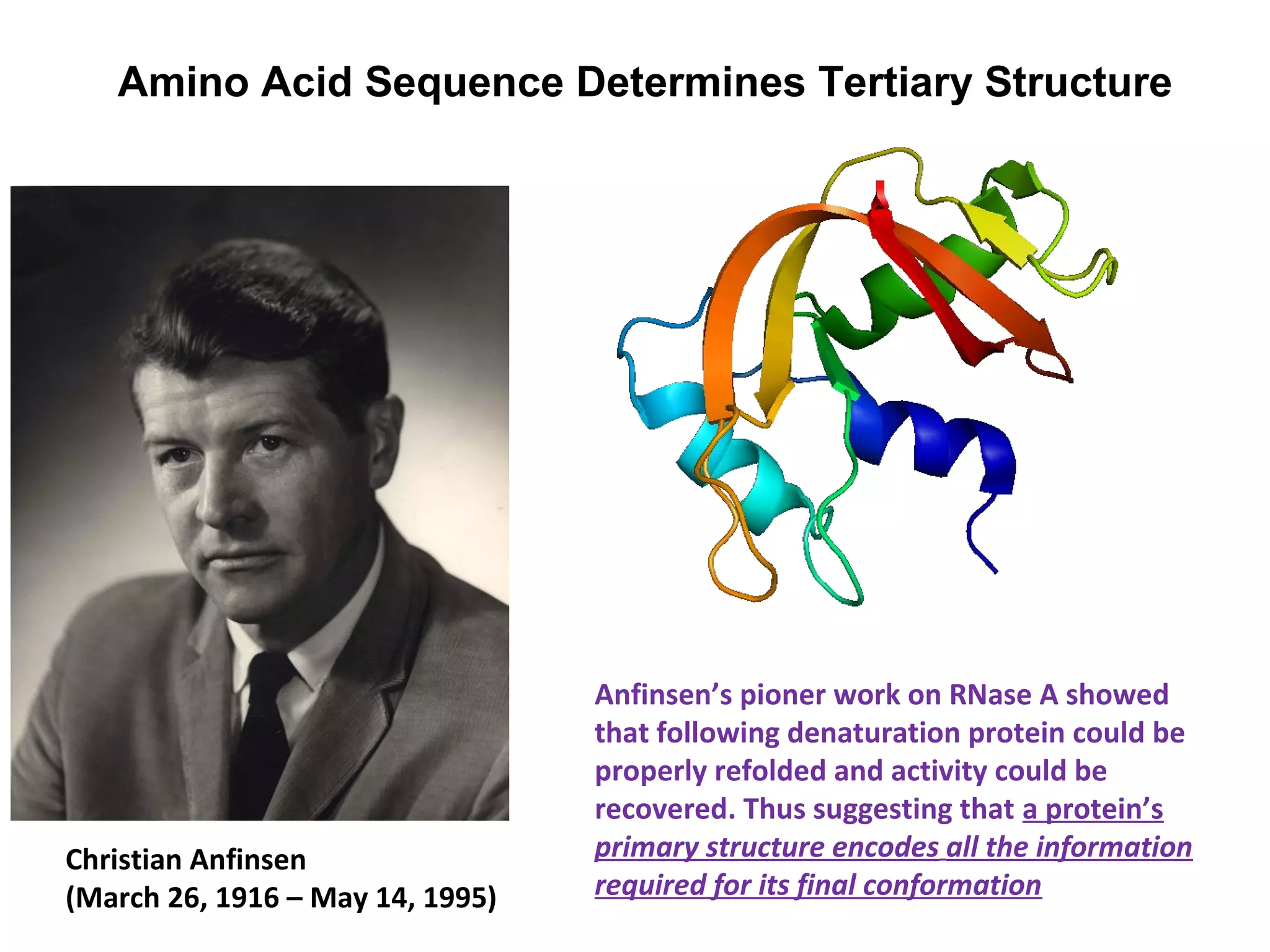
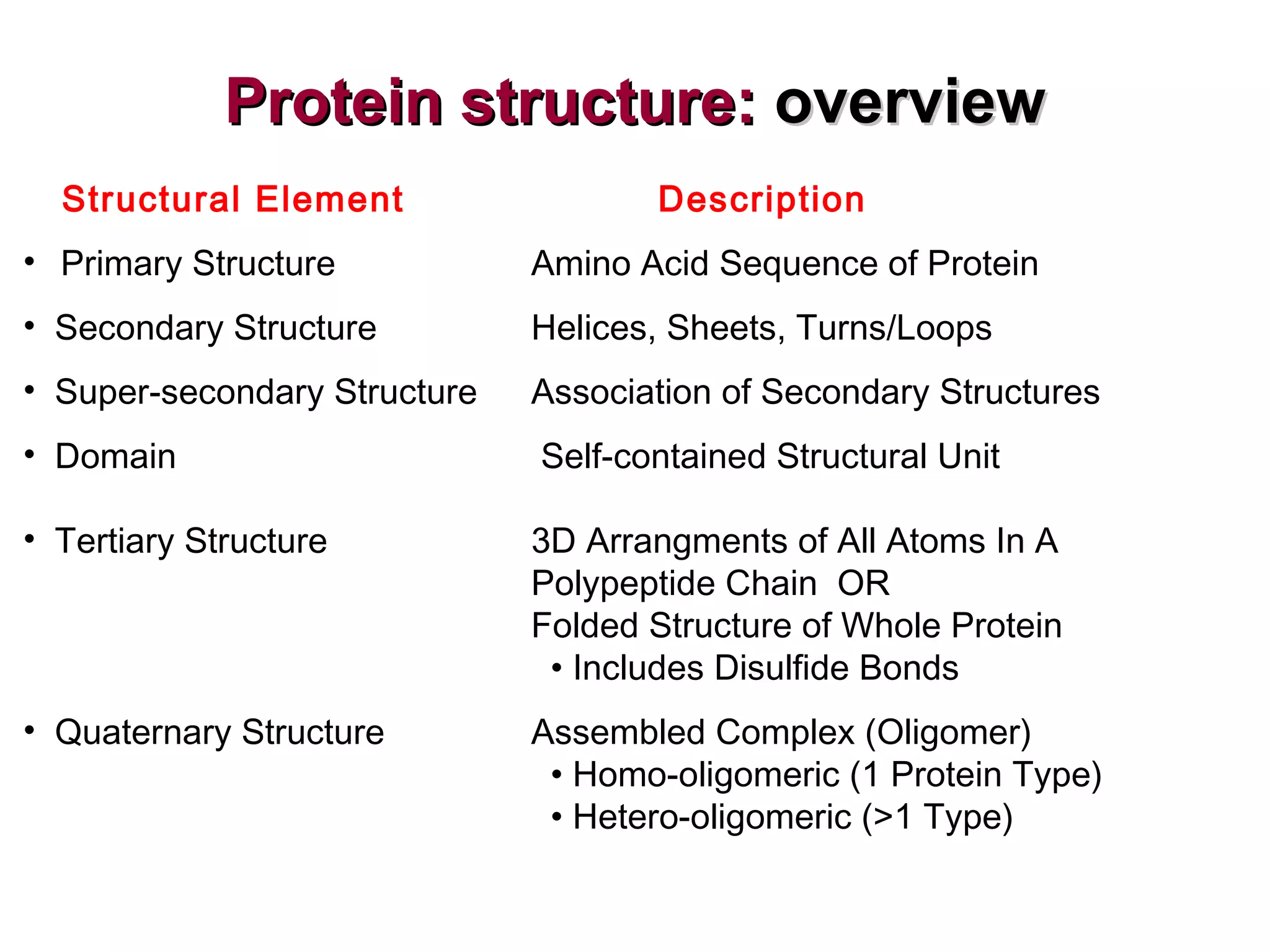
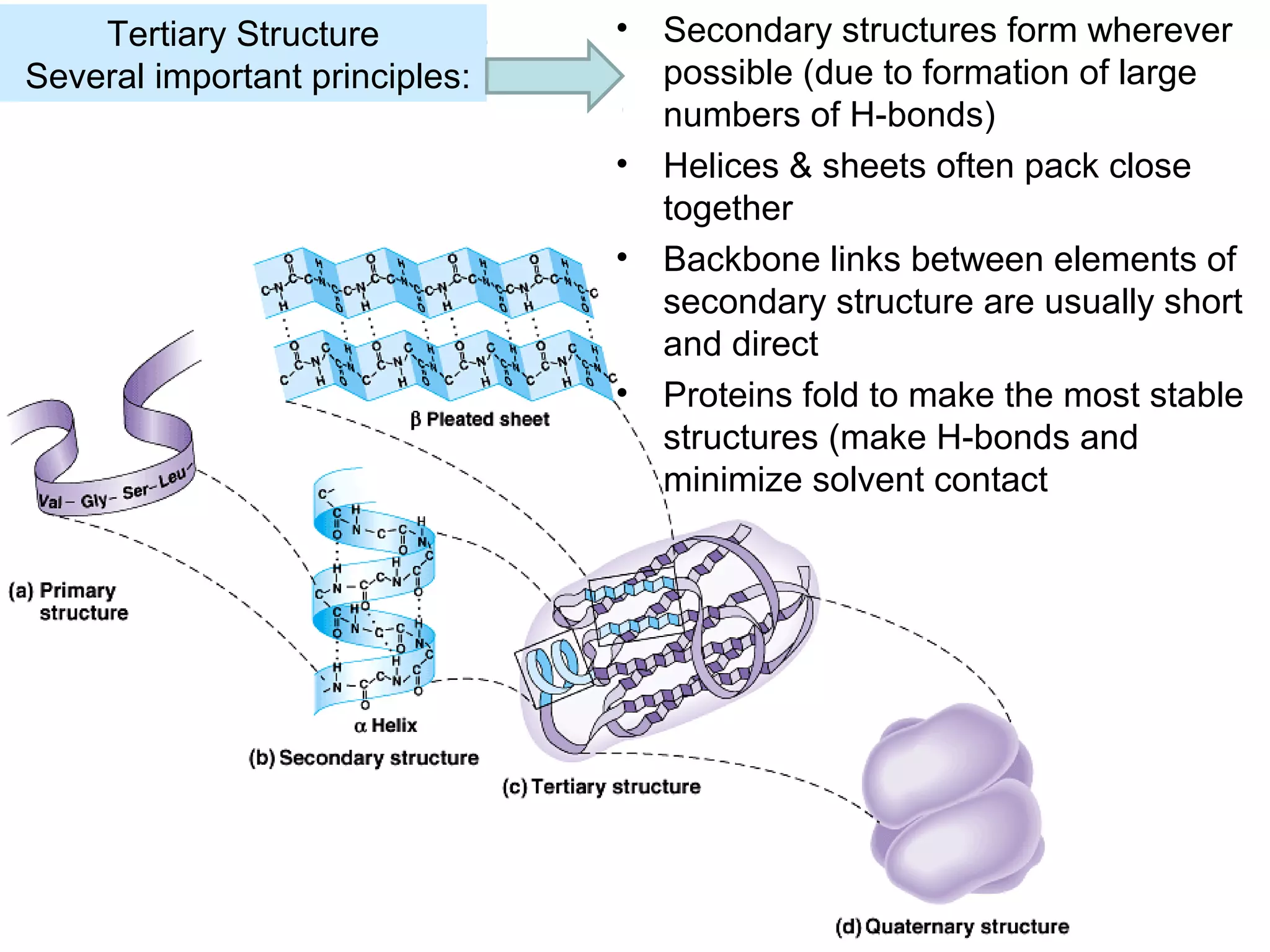
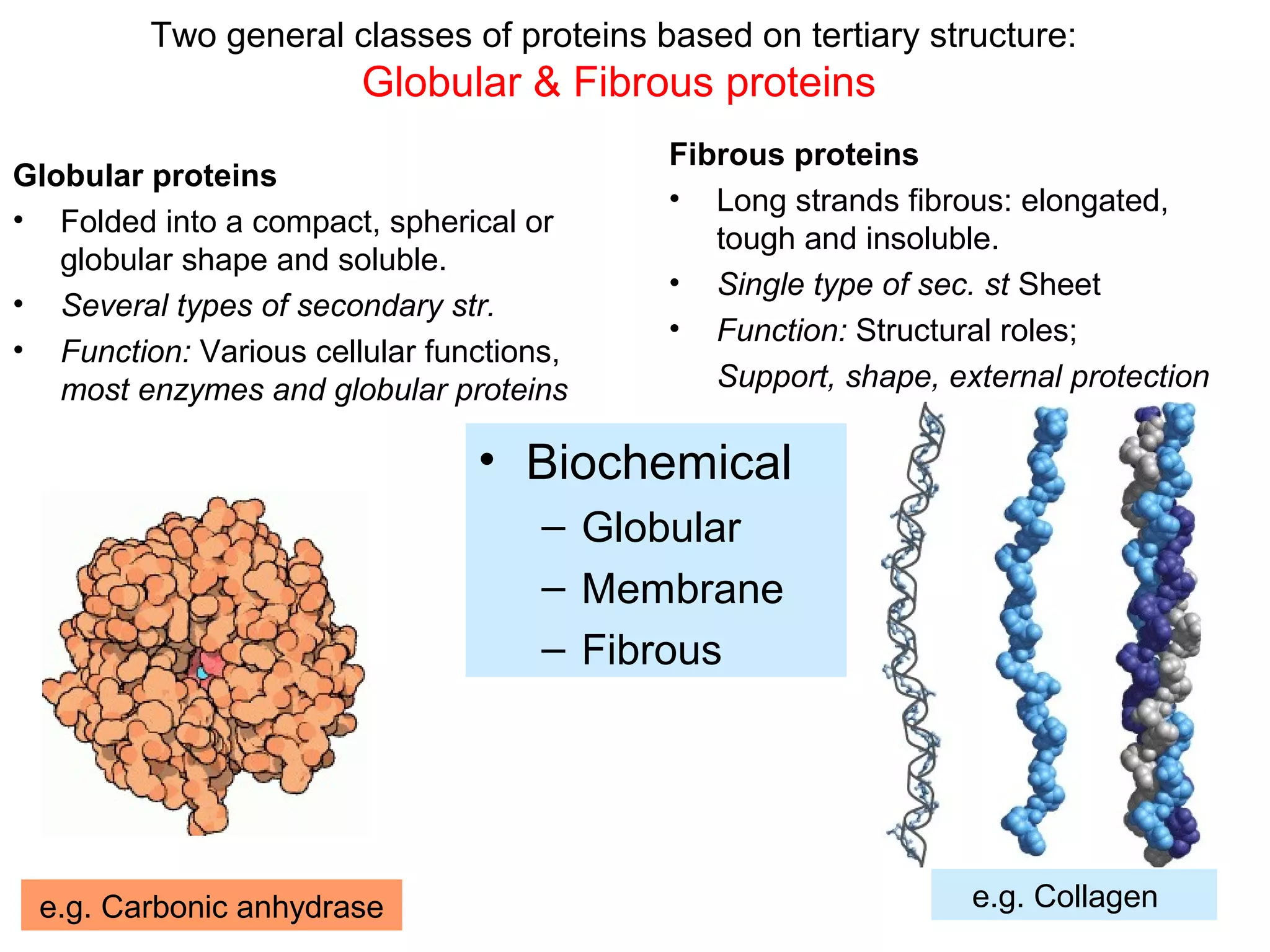
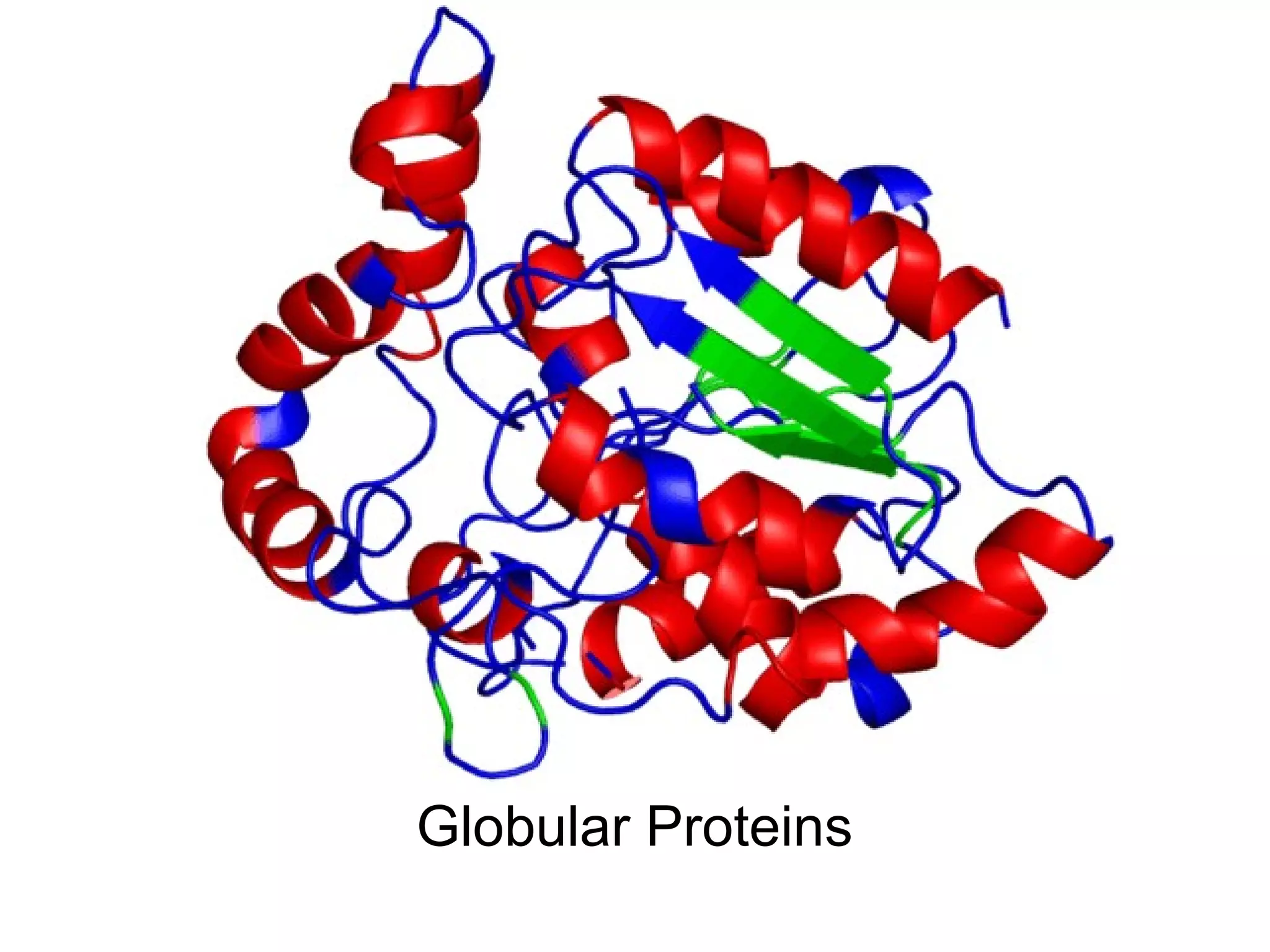
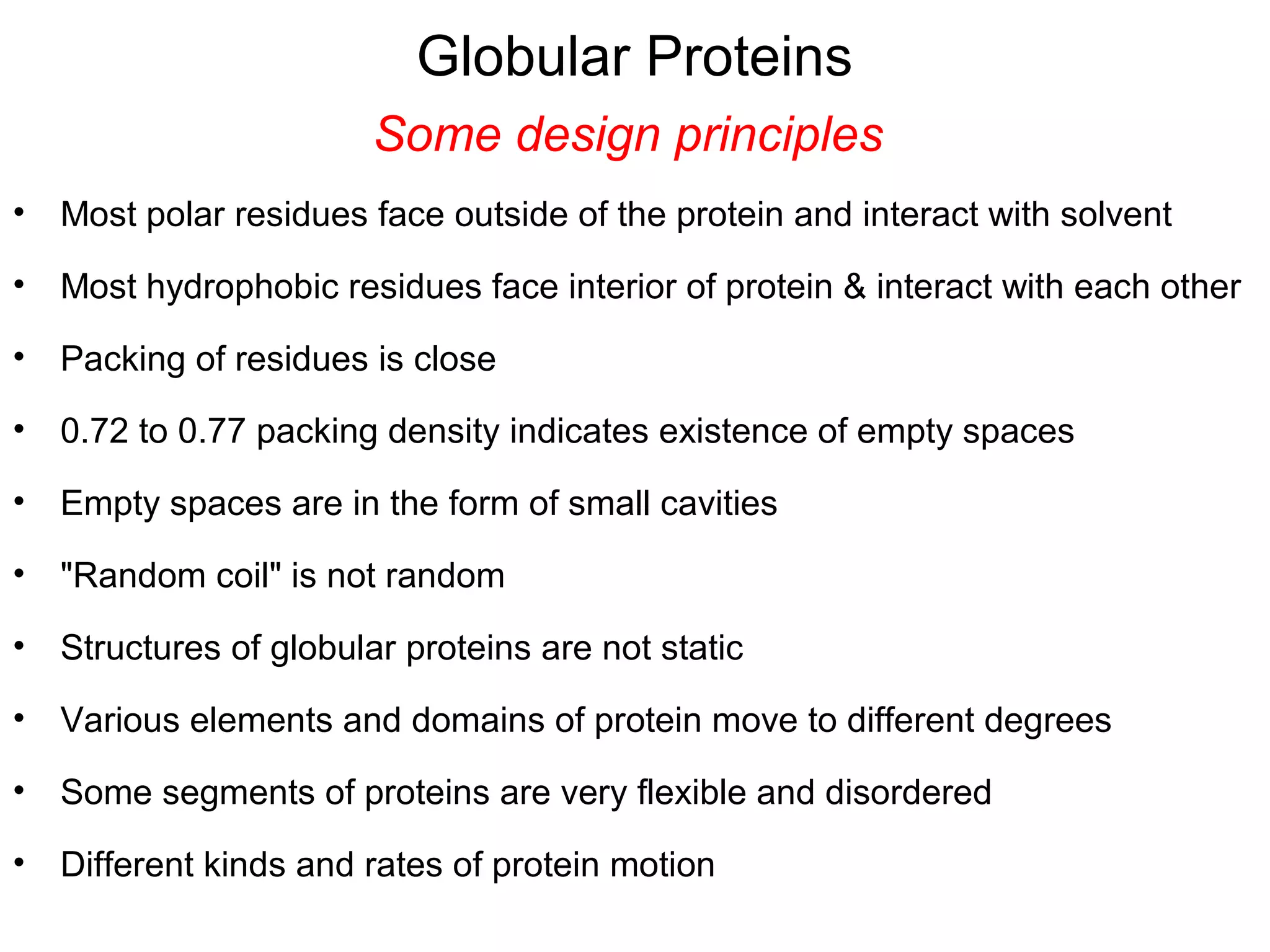
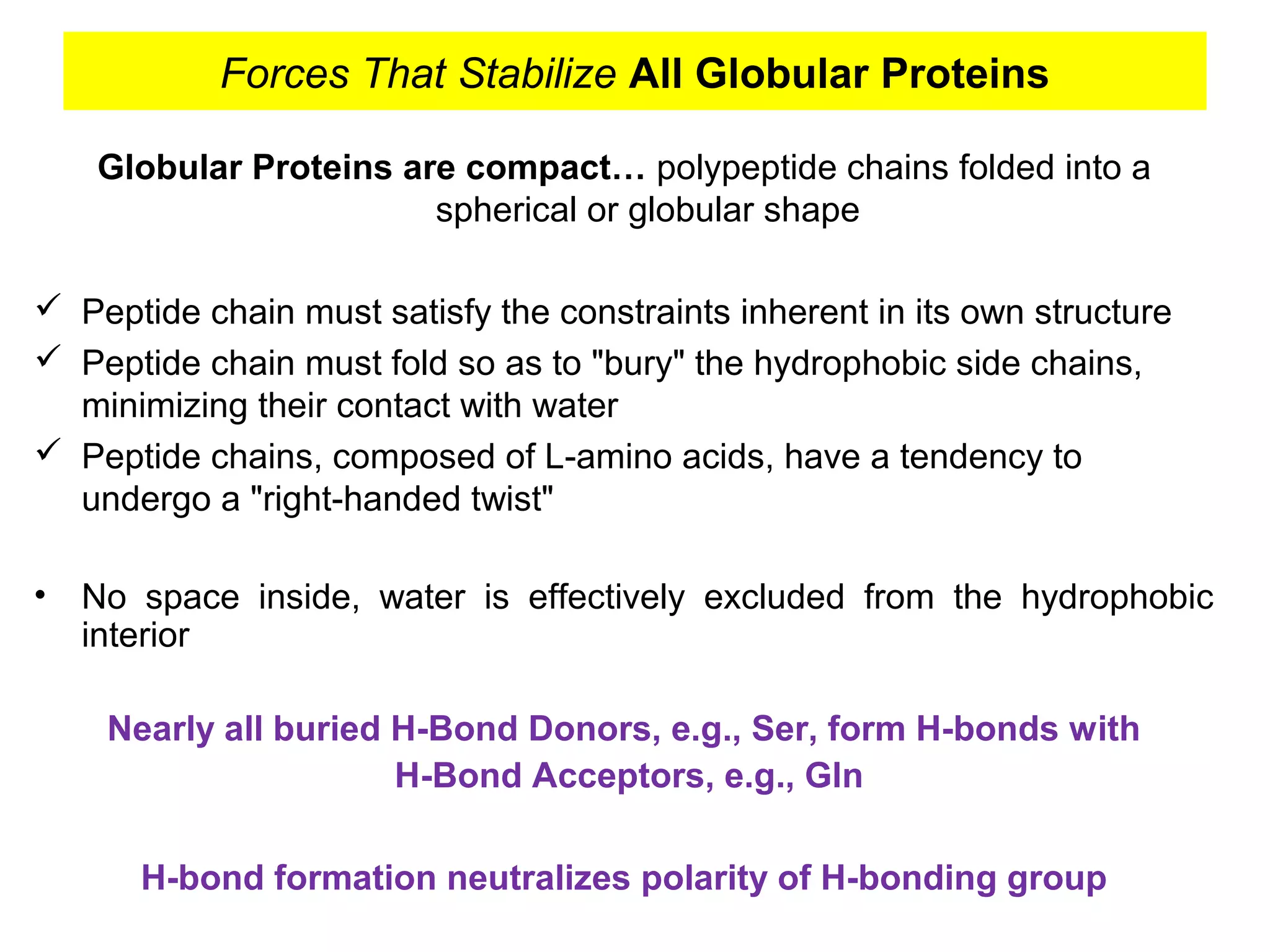
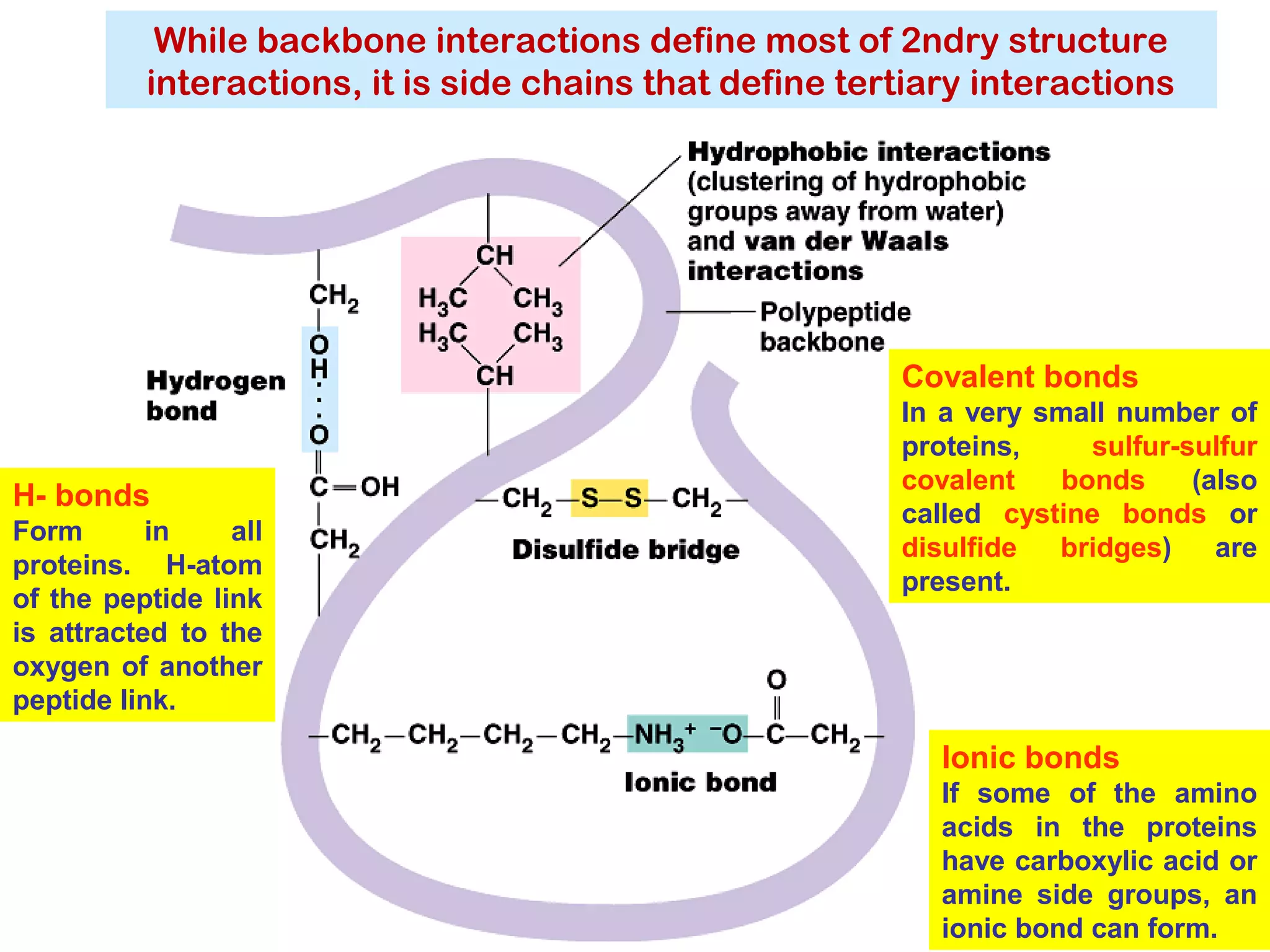
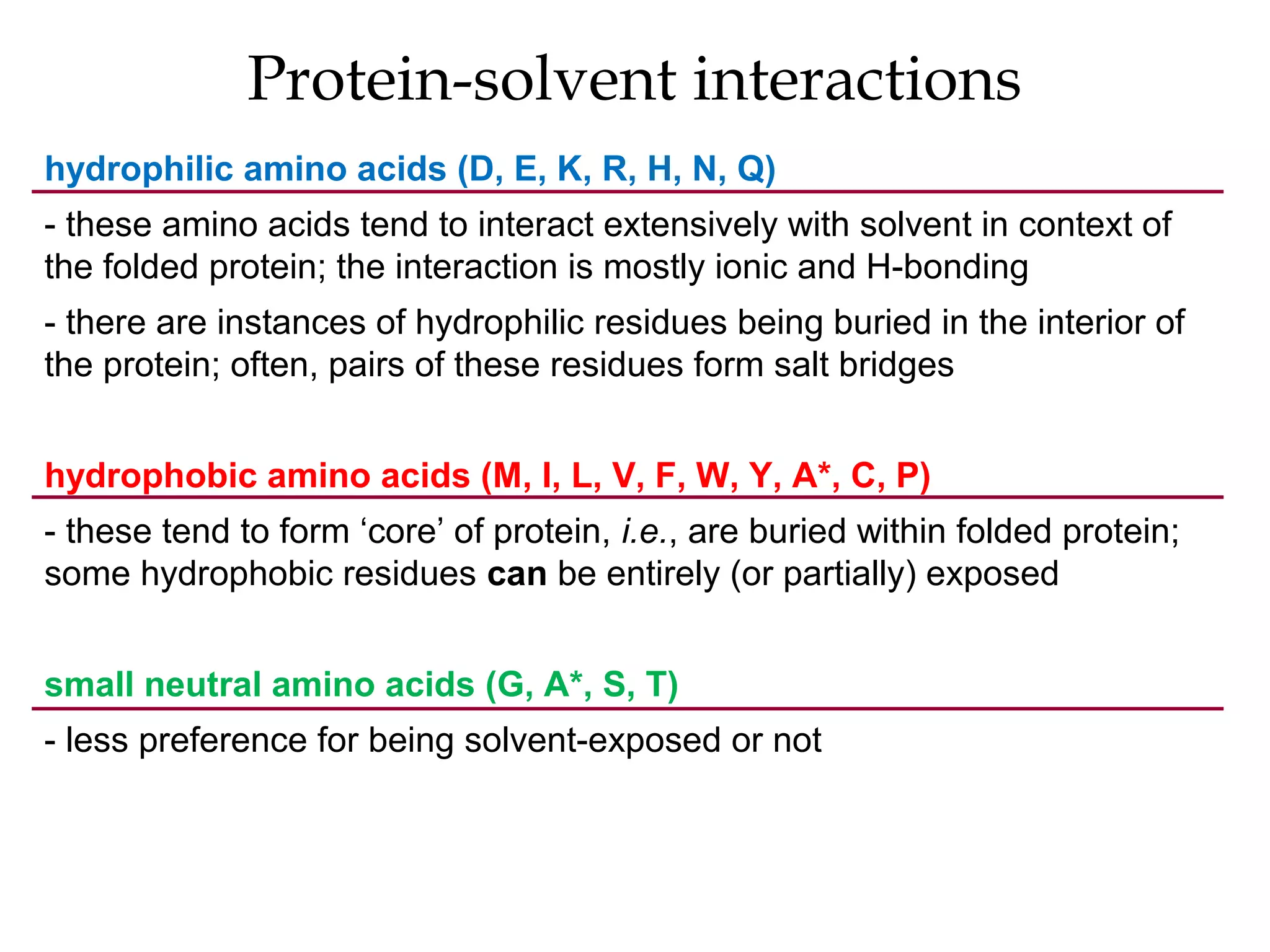
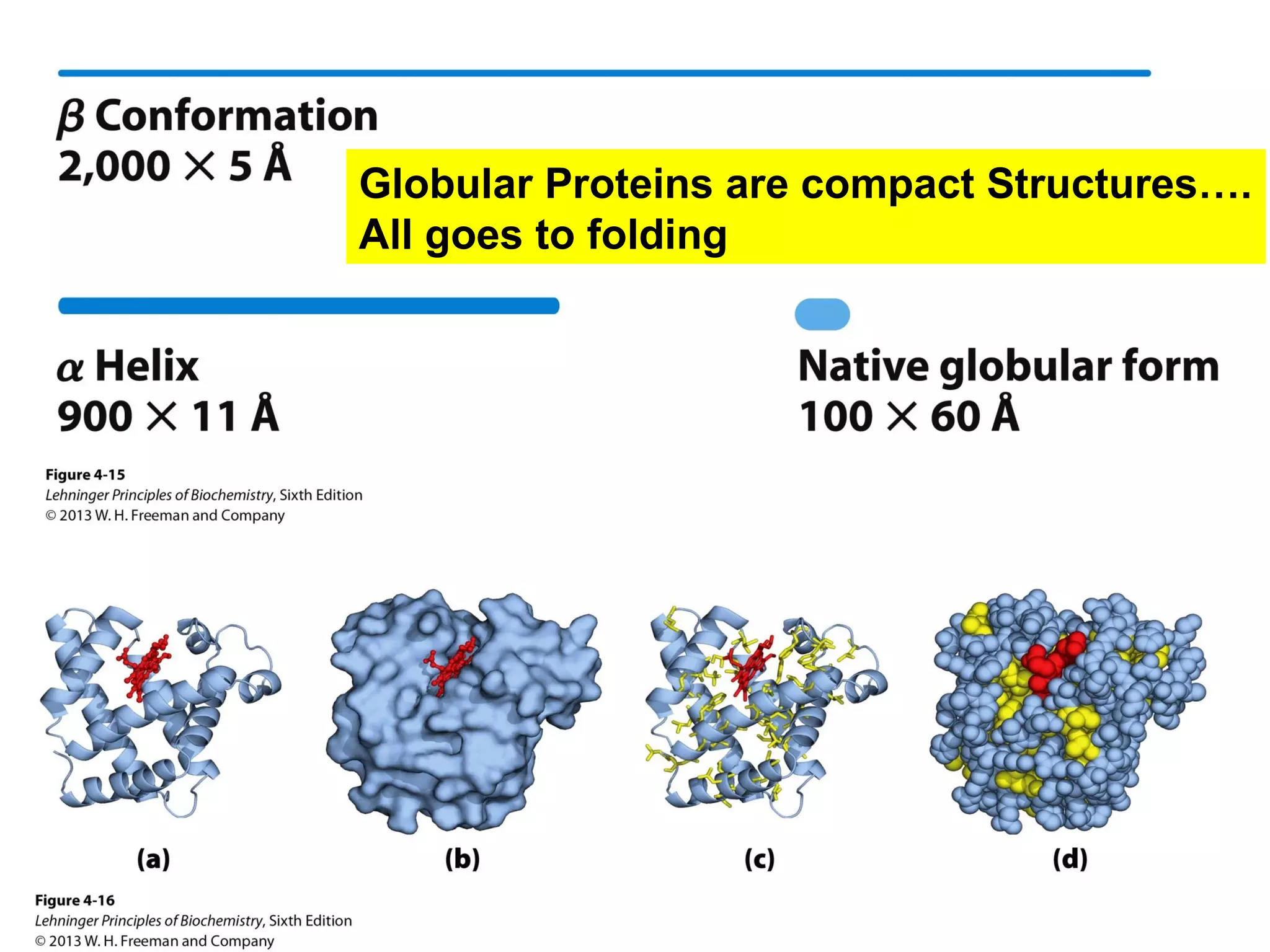
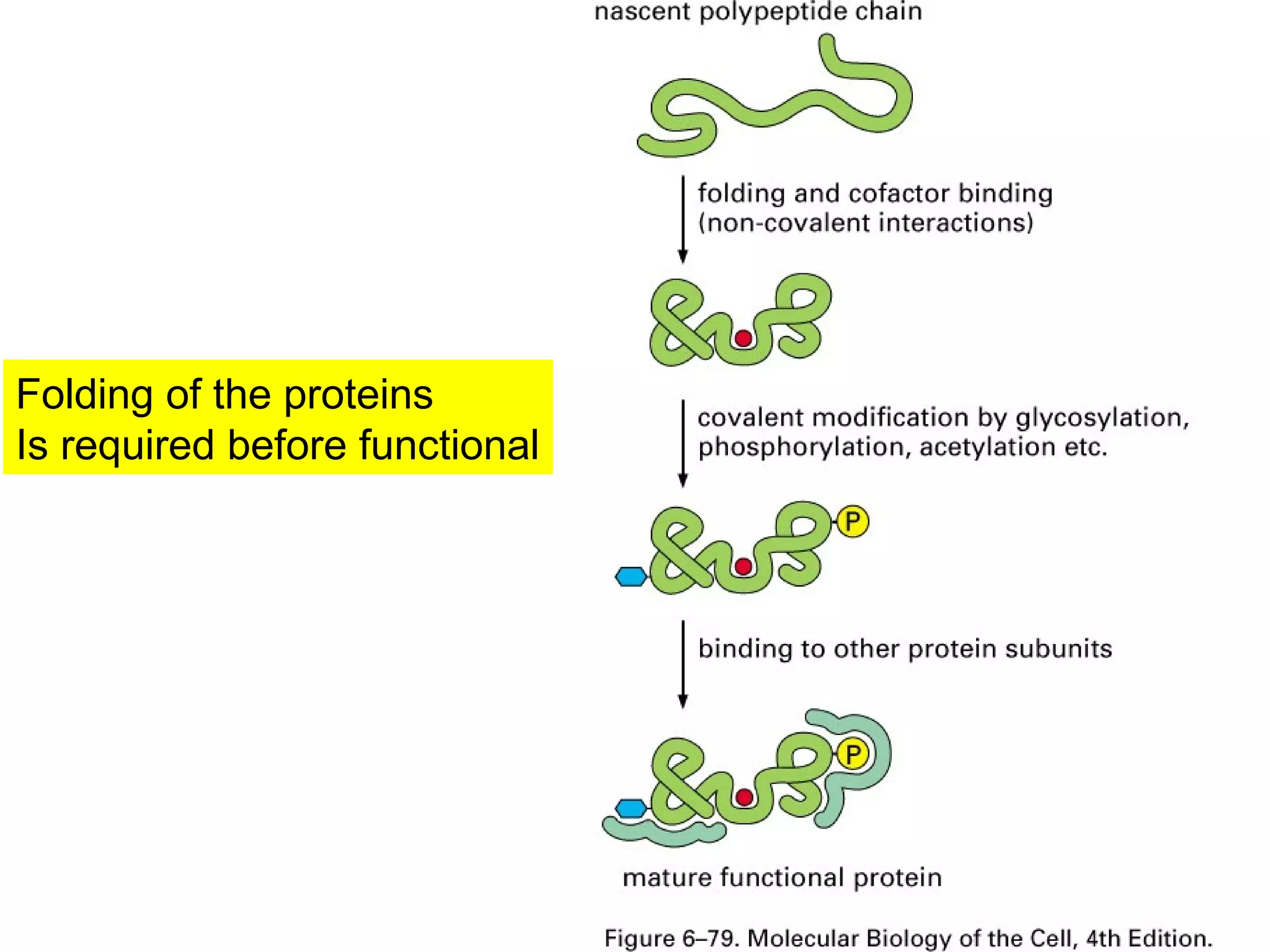
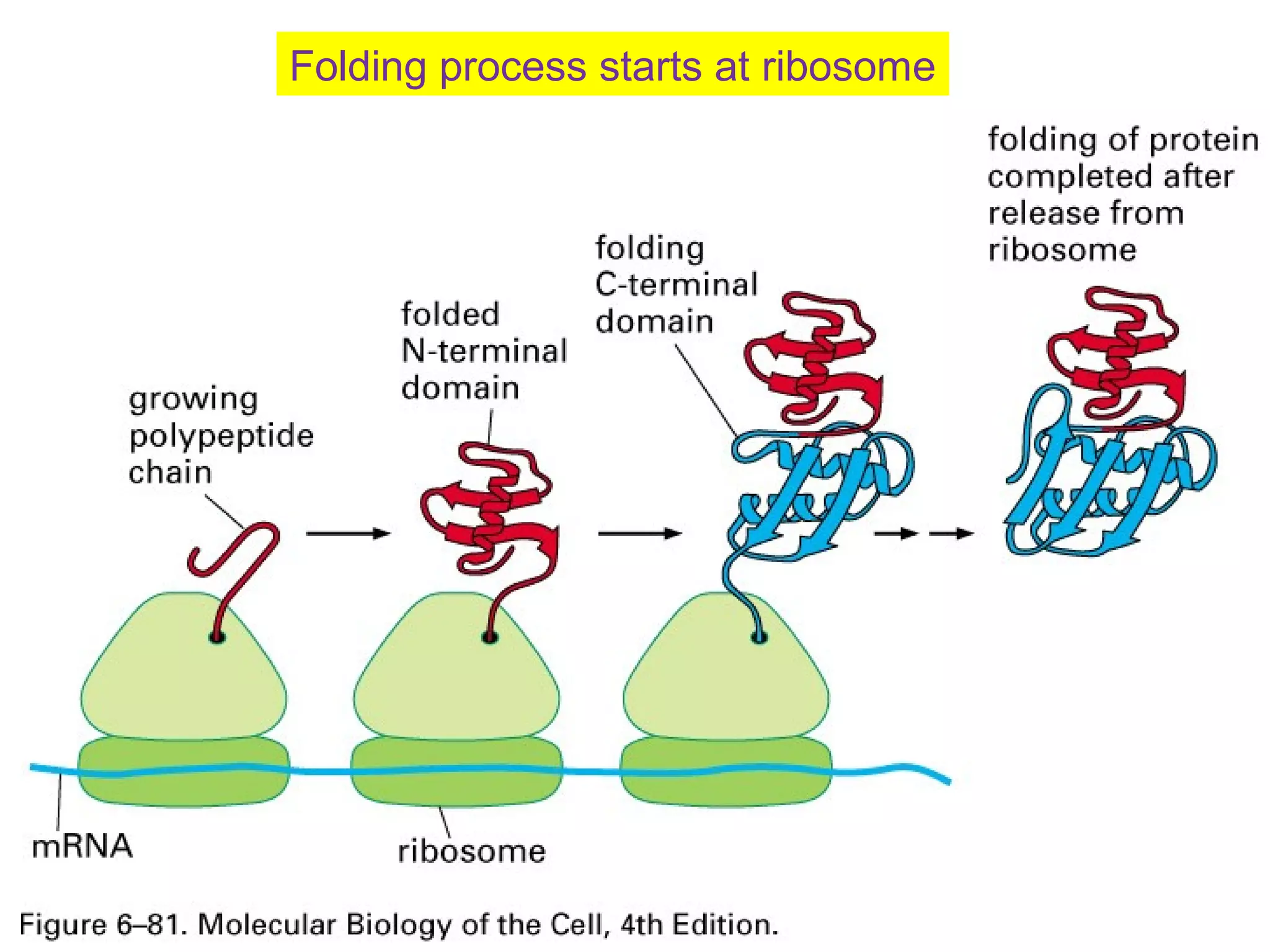
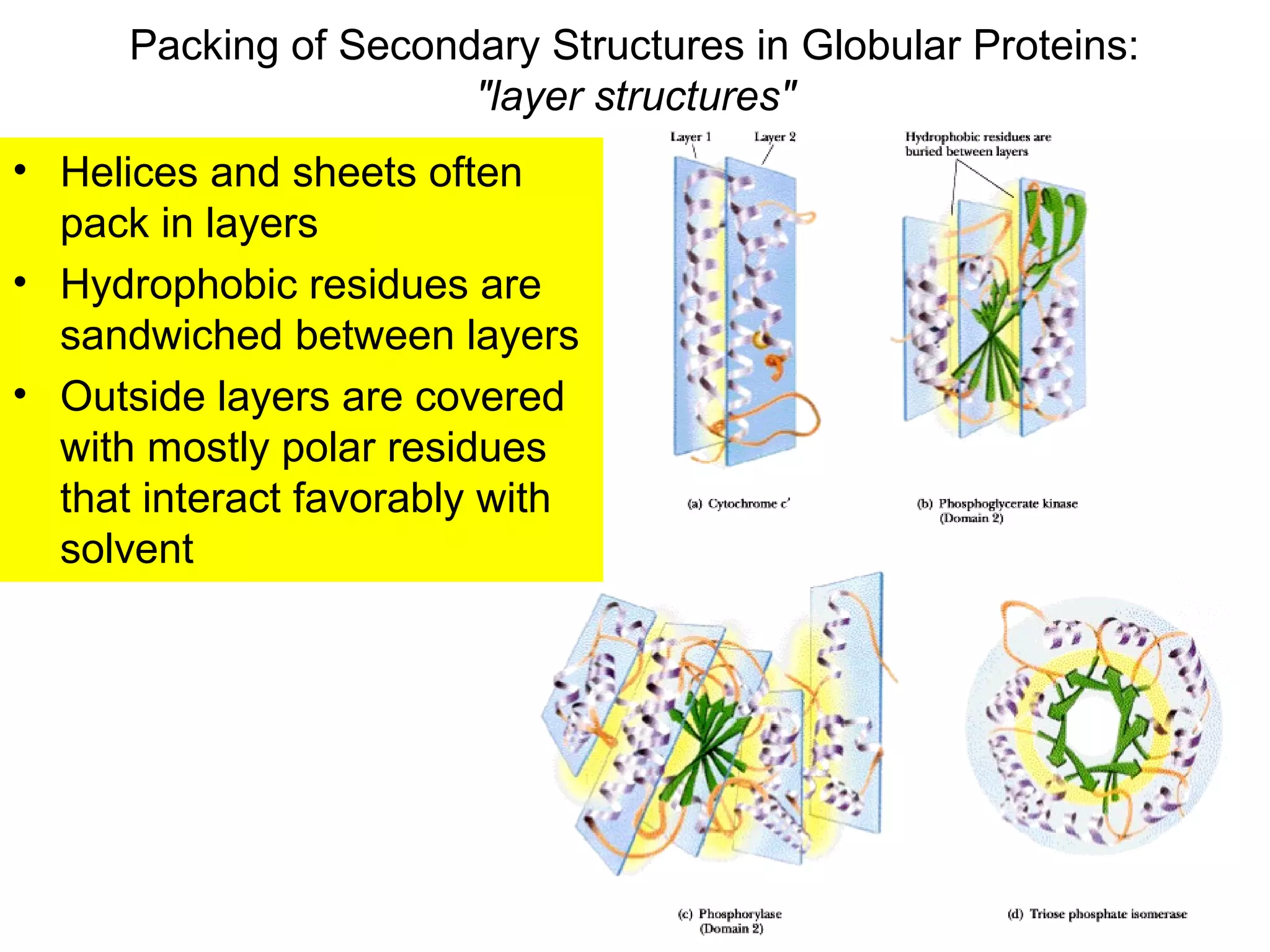
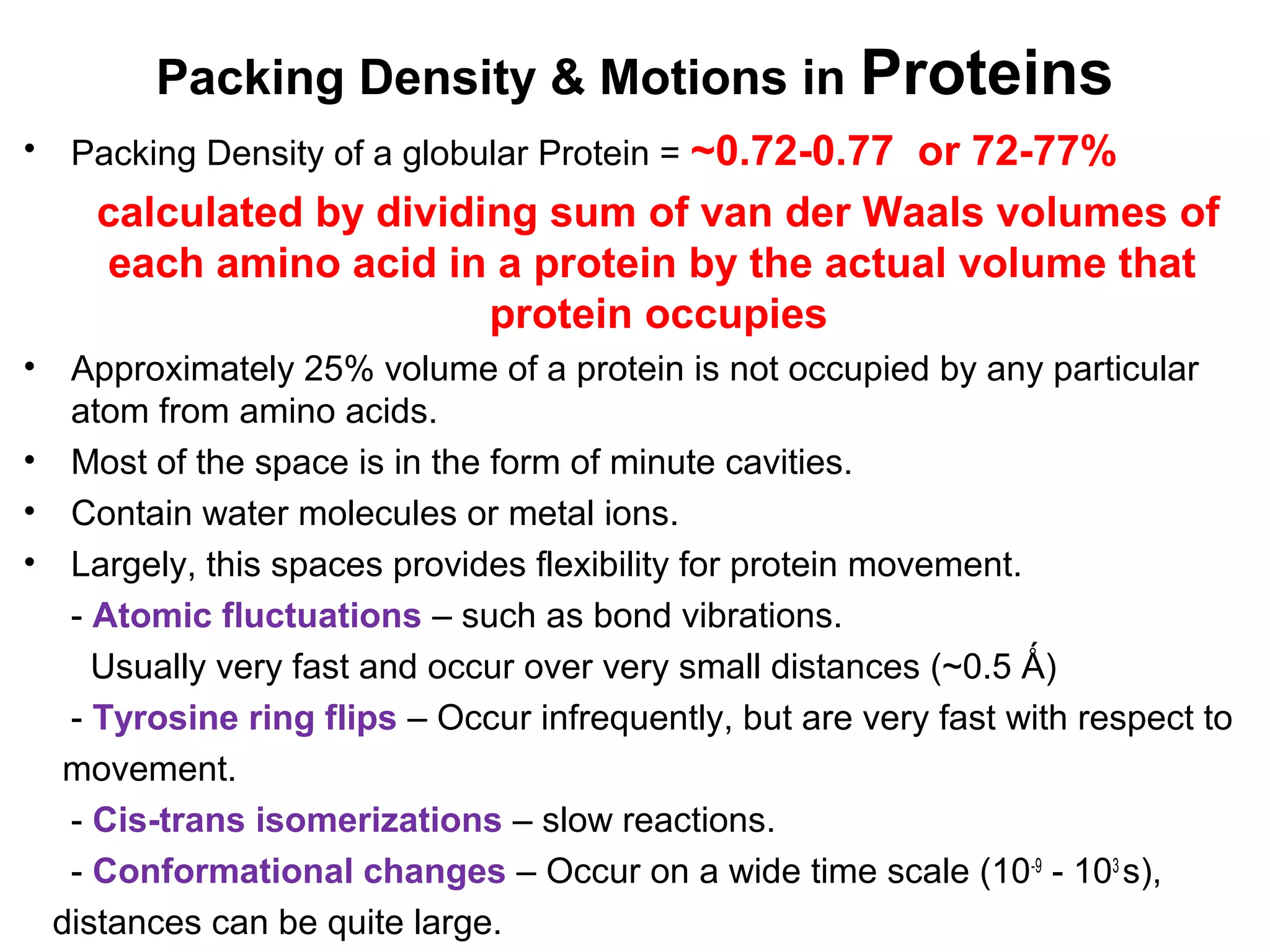
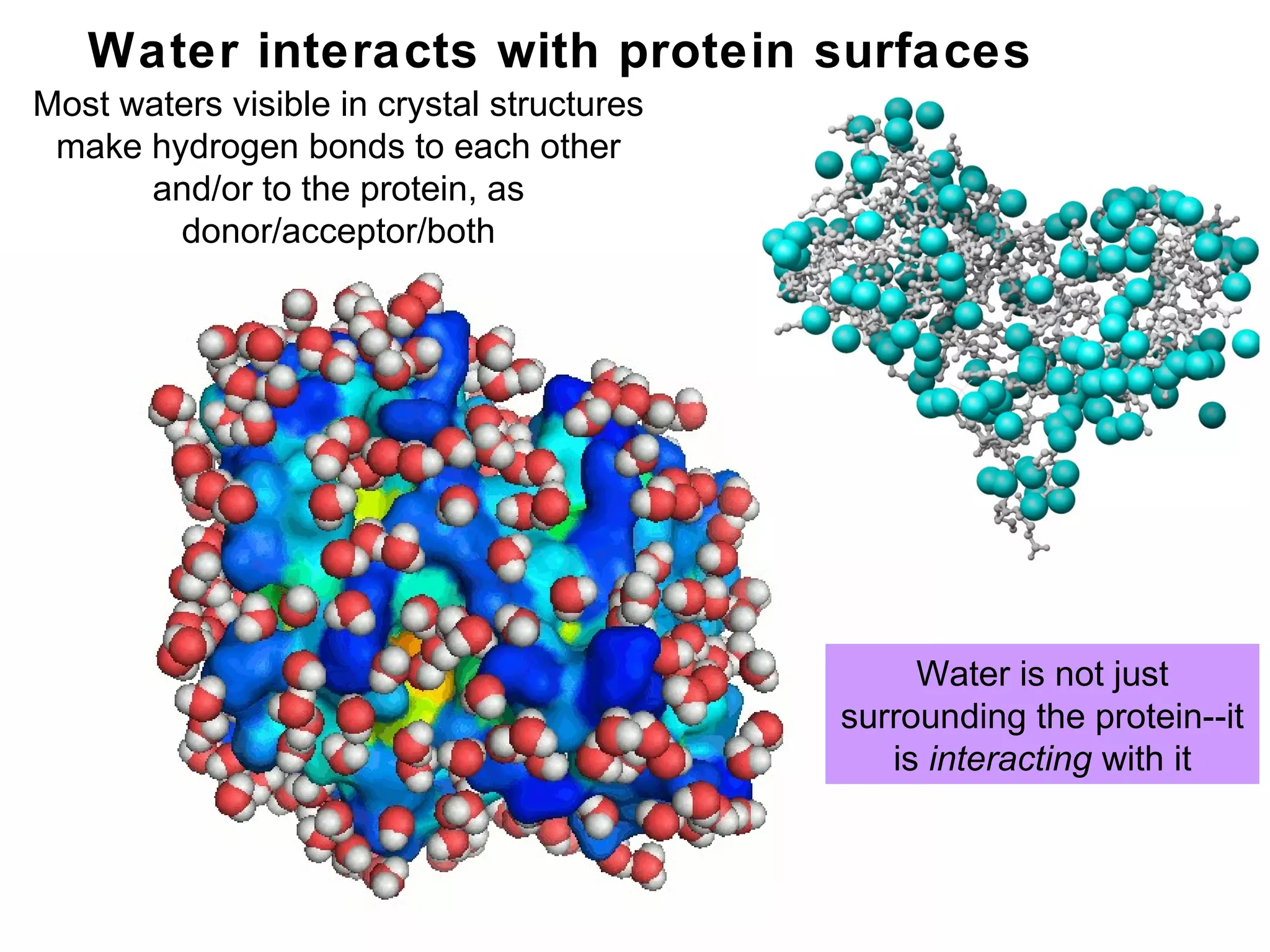
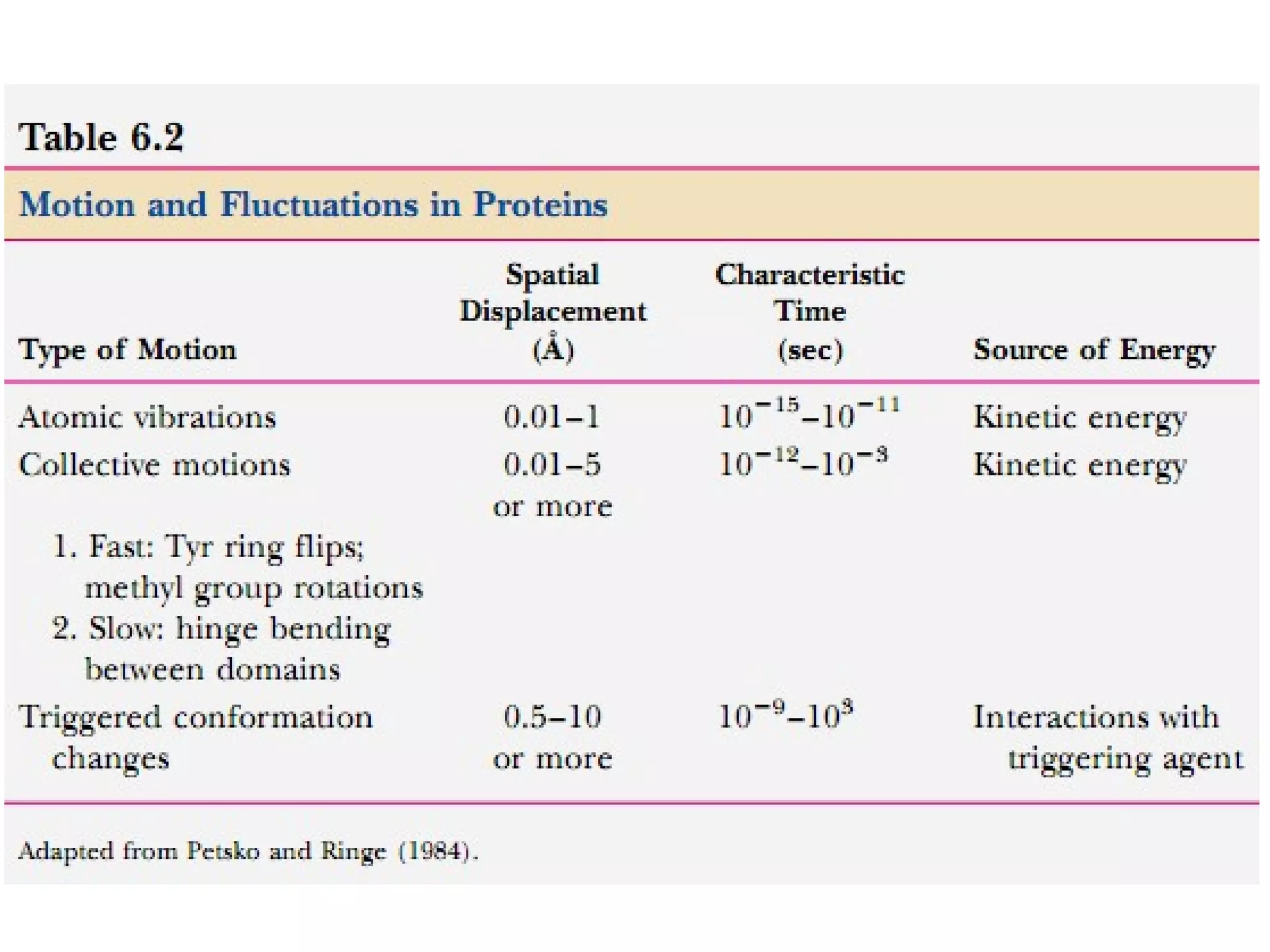
The document provides a comprehensive overview of protein tertiary structure, detailing its classification into globular and fibrous proteins, as well as the principles governing protein folding and stability. It discusses the hierarchical levels of protein structure, the interactions that stabilize proteins, and the importance of amino acid sequences in determining their conformation. Additionally, it highlights the impact of environmental factors and post-translational modifications on protein structure and functionality.


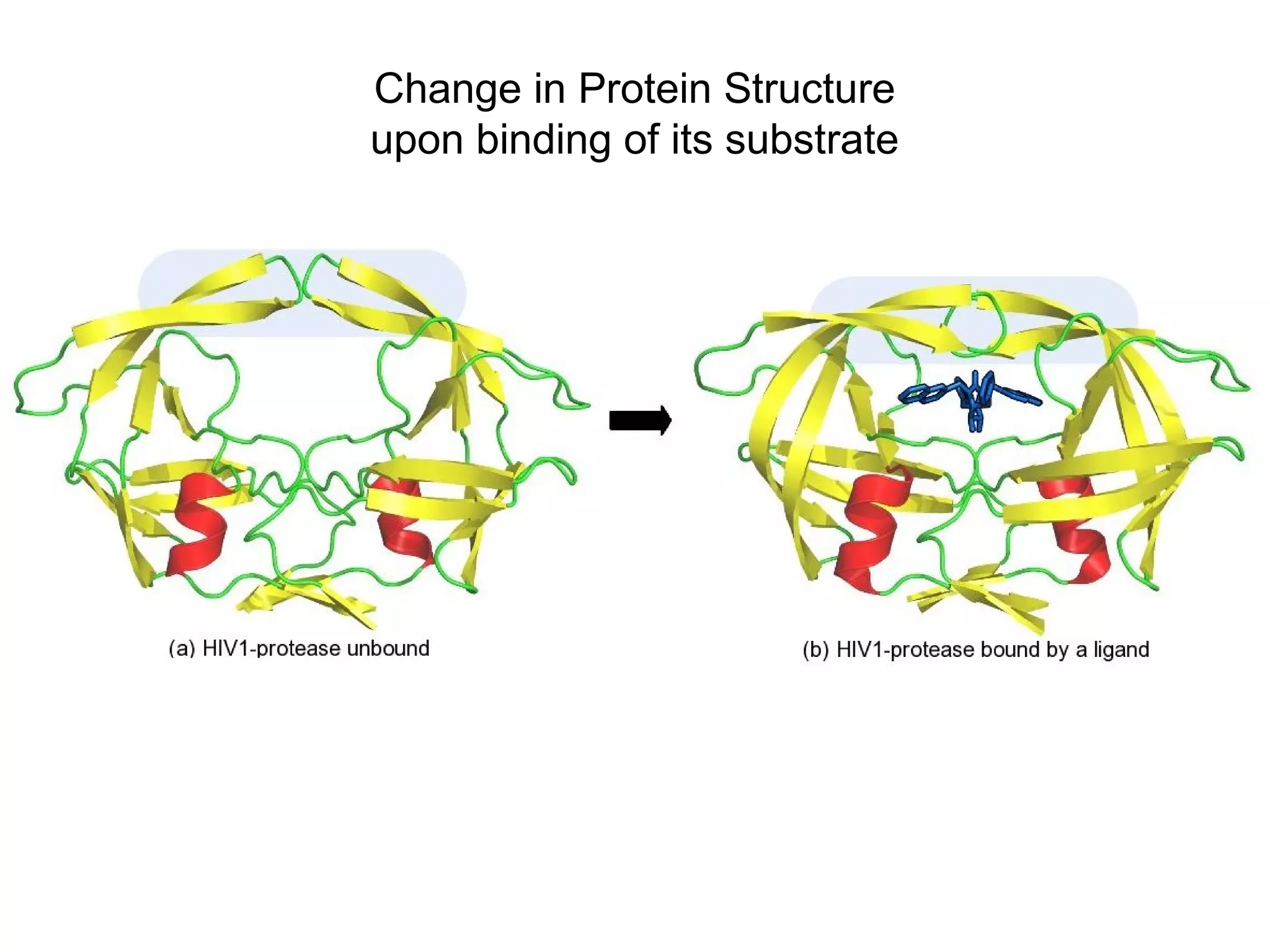
![General notion in enzymology
has been that substrate [on
which the enzyme acts] induces
shape change. We found that
this is not true. The enzyme, in
fact, changes conformations
without the substrate.
Image: Courtesy of Dorothee Kern/HHMI at
Brandeis University](https://image.slidesharecdn.com/8proetinstructuretertiary-190218092451/75/Proetin-Tertiary-Structure-23-2048.jpg)