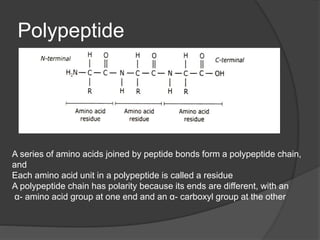
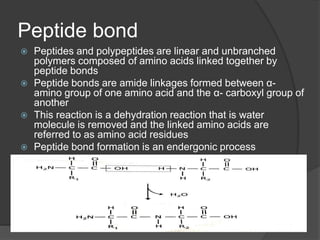
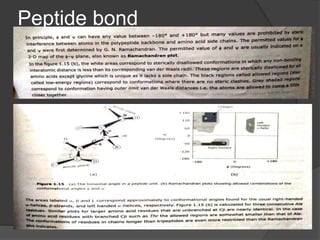
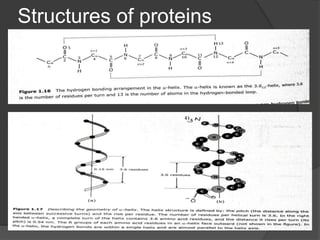
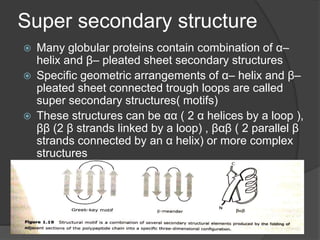
The document provides an overview of peptides and proteins, explaining their structures, including primary, secondary, tertiary, and quaternary formations. It describes the formation and significance of peptide bonds, the roles of various peptides and proteins in biological processes, and factors affecting protein solubility and denaturation. Additionally, it outlines the impact of external agents on protein stability and solubility, including pH, ionic strength, and temperature.