Polymerase Chain Reaction (PCR) is a technique that amplifies a specific DNA sequence. It involves denaturing DNA, annealing primers to the DNA, and extending the primers to replicate the DNA. Through repeated cycles of heating and cooling, the targeted DNA sequence is exponentially amplified, allowing it to be detected and manipulated. PCR is used to identify infectious organisms like viruses, detect genetic mutations, and replicate DNA for further analysis. Careful primer design is important for PCR to specifically amplify the desired DNA sequence.







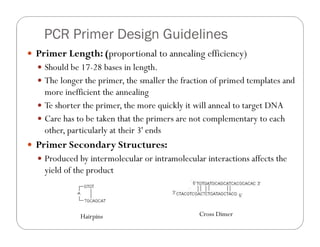

![ The GC content of the sequence gives a fair indication of the
primerTm
The formula for primerTm calculation:
Tm = 4(G + C) + 2(A +T)=°C
[Base composition should be 50-60% (G+C)]
Presence of G or C bases within the last five bases from the 3'
end, known as the GC clamp
Promote specific binding at the 3' end due to the stronger bonding
of G and C bases.
More than 3 G's or C's should be avoided in the last 5 bases at
the 3' end of the primer (promote mispriming)](https://image.slidesharecdn.com/pcrandprimerdesign604-230611034504-10d9745b/85/PCR-and-Primer-design-604-pdf-11-320.jpg)