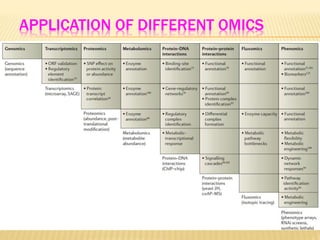
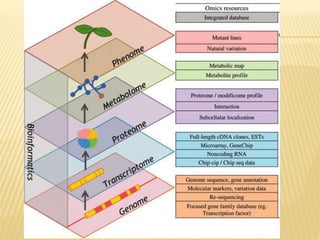
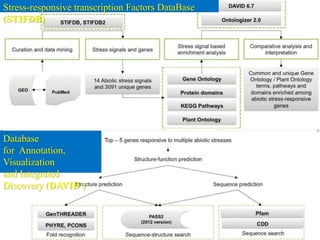
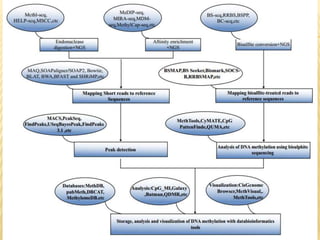
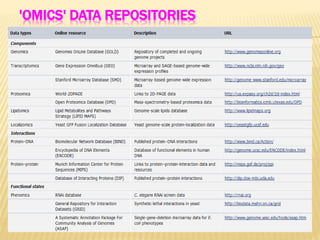
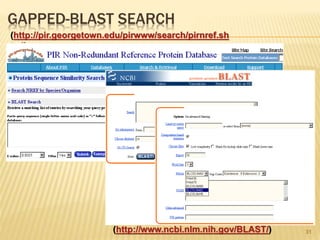
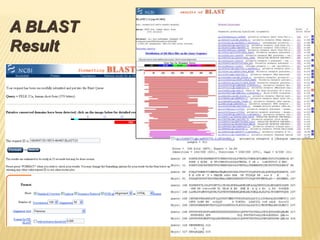
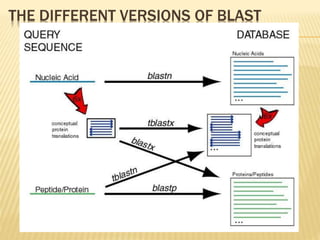
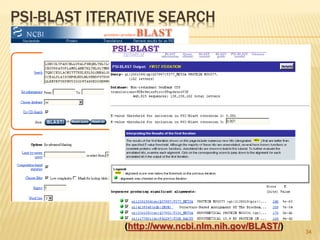
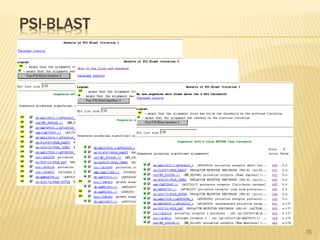
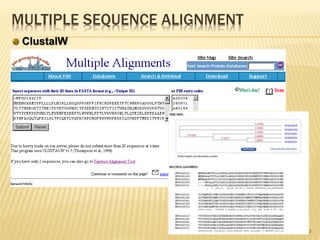
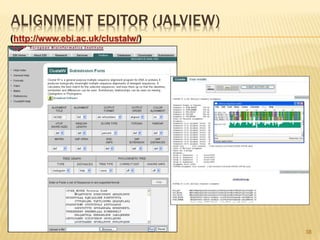
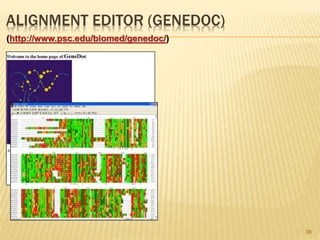
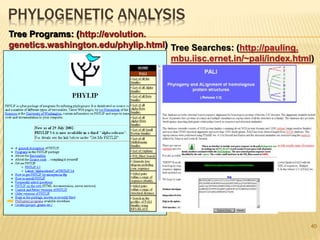
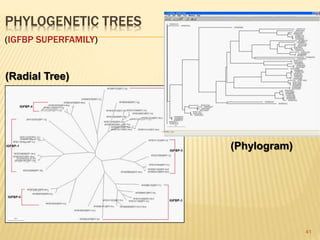
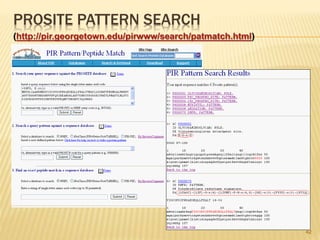
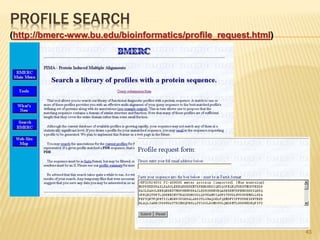
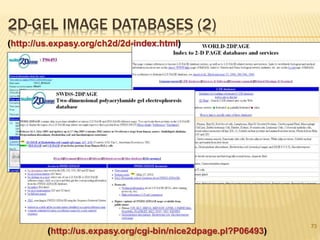
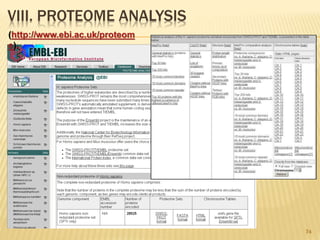
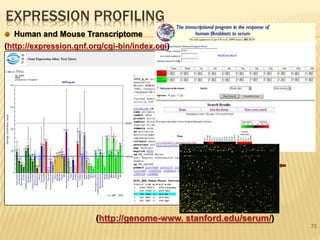
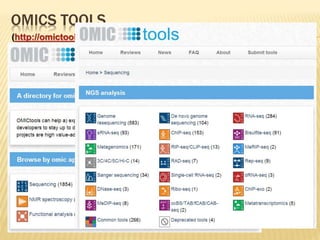
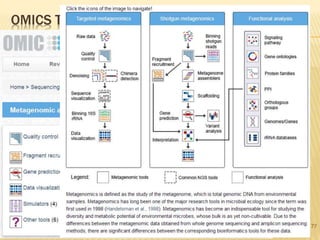
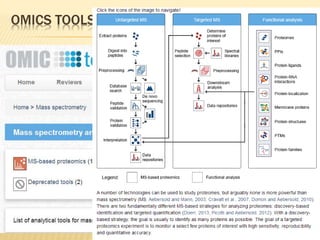
The document discusses various "omics" fields of study including genomics, proteomics, metabolomics, and others. It provides definitions and descriptions of each type of omics, focusing on the large sets of biological molecules they each study such as genomes, proteomes, metabolomes, etc. It explains that omics fields examine biological data on a large scale and provide insights into biological processes, functions, and interactions on a systems-wide level.









![COMPUTATIONAL GENOMICS
Computational genomics (often referred to as
Computational Genetics) refers to the use of
computational and statistical analysis to decipher
biology from genome sequences and related
data,[1] including both DNA and RNA sequence as
well as other "post-genomic" data (i.e. experimental
data obtained with technologies that require the
genome sequence, such as genomic DNA
microarrays). These, in combination with
computational and statistical approaches to
understanding the function of the genes and
statistical association analysis, this field is also often
referred to as Computational and Statistical
Genetics/genomics.
13](https://image.slidesharecdn.com/omicsera-140630223756-phpapp02/85/Omics-era-13-320.jpg)

![PATHOGENOMICS
Pathogen infections are among the leading causes of
infirmity and mortality among humans and other animals
in the world.[1] Until recently, it has been difficult to
compile information to understand the generation of
pathogen virulence factors as well as pathogen
behaviour in a host environment. The study of
Pathogenomics attempts to utilize genomic and
metagenomics data gathered from high through-put
technologies (e.g. sequencing or DNA microarrays), to
understand microbe diversity and interaction as well as
host-microbe interactions involved in disease states.
The bulk of pathogenomics research concerns itself with
pathogens that affect human health; however, studies
also exist for plant and animal infecting microbes.
17](https://image.slidesharecdn.com/omicsera-140630223756-phpapp02/85/Omics-era-17-320.jpg)
![REGENOMICS
Regenomics represents the merger of two fields of
scientific endeavor: Regenerative medicine[1] and
genomics.[2][3][4] New technologies to reprogram aged
somatic cells back to pluripotency and to restore
telomere length are currently used in research in
regenerative medicine,[5] though FDA-approved cellular
therapies using reprogrammed cells are currently not
available in the United States.[6] The culture and
banking of somatic cells also allows the parallel
sequencing of their nuclear DNA to provide individuals
with potentially valuable information for guiding them in
lifestyle choices, but also one day, potentially in
preventative strategies where cell types are made in
advance for high risk categories of disease, i.e.
preparing cardiac progenitor cells for individuals at high
risk for heart disease.
18](https://image.slidesharecdn.com/omicsera-140630223756-phpapp02/85/Omics-era-18-320.jpg)
![PERSONAL GENOMICS
Personal genomics is the branch of genomics concerned with
the sequencing and analysis of the genome of an individual.
The genotyping stage employs different techniques, including
single-nucleotide polymorphism (SNP) analysis chips (typically
0.02% of the genome), or partial or full genome sequencing.
Once the genotypes are known, the individual's genotype can
be compared with the published literature to determine
likelihood of trait expression and disease risk.
Use of personal genomics in predictive and precision
medicine[edit]
Predictive medicine is the use of the information produced by
personal genomics techniques when deciding what medical
treatments are appropriate for a particular individual. Precision
medicine is focused on "a new taxonomy of human disease
based on molecular biology“.
19](https://image.slidesharecdn.com/omicsera-140630223756-phpapp02/85/Omics-era-19-320.jpg)