This document provides an overview of next generation sequencing (NGS), including its history, key methods, applications and challenges. It discusses that NGS allows for massively parallel sequencing of thousands of DNA molecules simultaneously, providing exact sequences of genes. Some main NGS platforms and their sequencing methods are described, such as Illumina sequencing by synthesis. Applications of NGS discussed include cancer research, pharmacogenetics and preimplantation genetic diagnosis. Challenges associated with NGS like poor quality FFPE samples and large data analysis workflows are also outlined.

![● ABM. n.d. Next Generation Sequencing (NGS) - An Introduction, viewed 25 May 2019,
<https://www.abmgood.com/marketing/knowledge_base/next_generation_sequencing_introduction.php>
● Ambardar, S., Gupta, R., Trakroo, D., Lal, R. and Vakhlu, J. 2016. ‘High Throughput Sequencing: An Overview of Sequencing
Chemistry. Indian Journal of Microbiology’, [online] 56(4), pp.394-404. Available at:
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5061697/ [Accessed 27 May 2019].
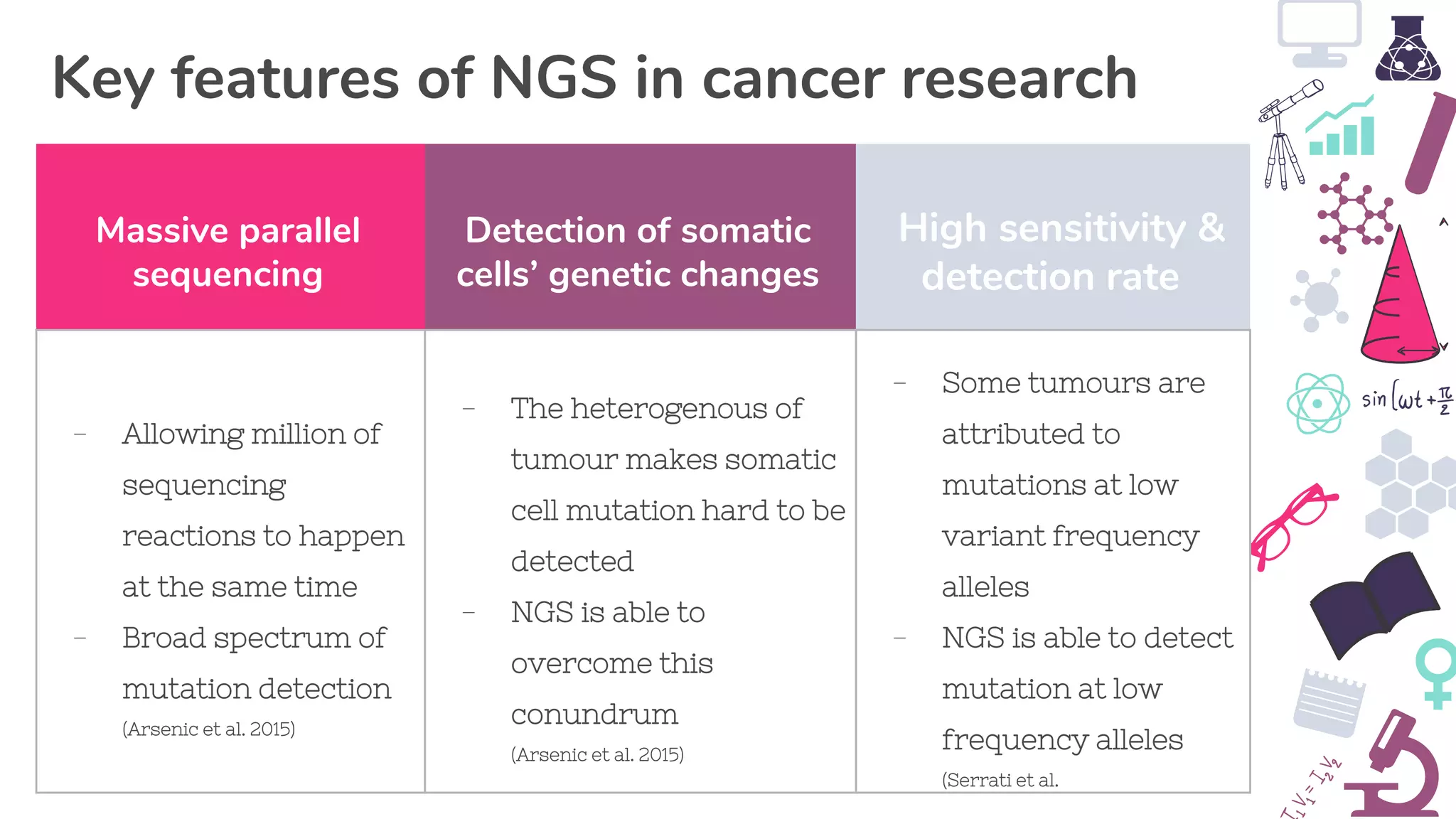
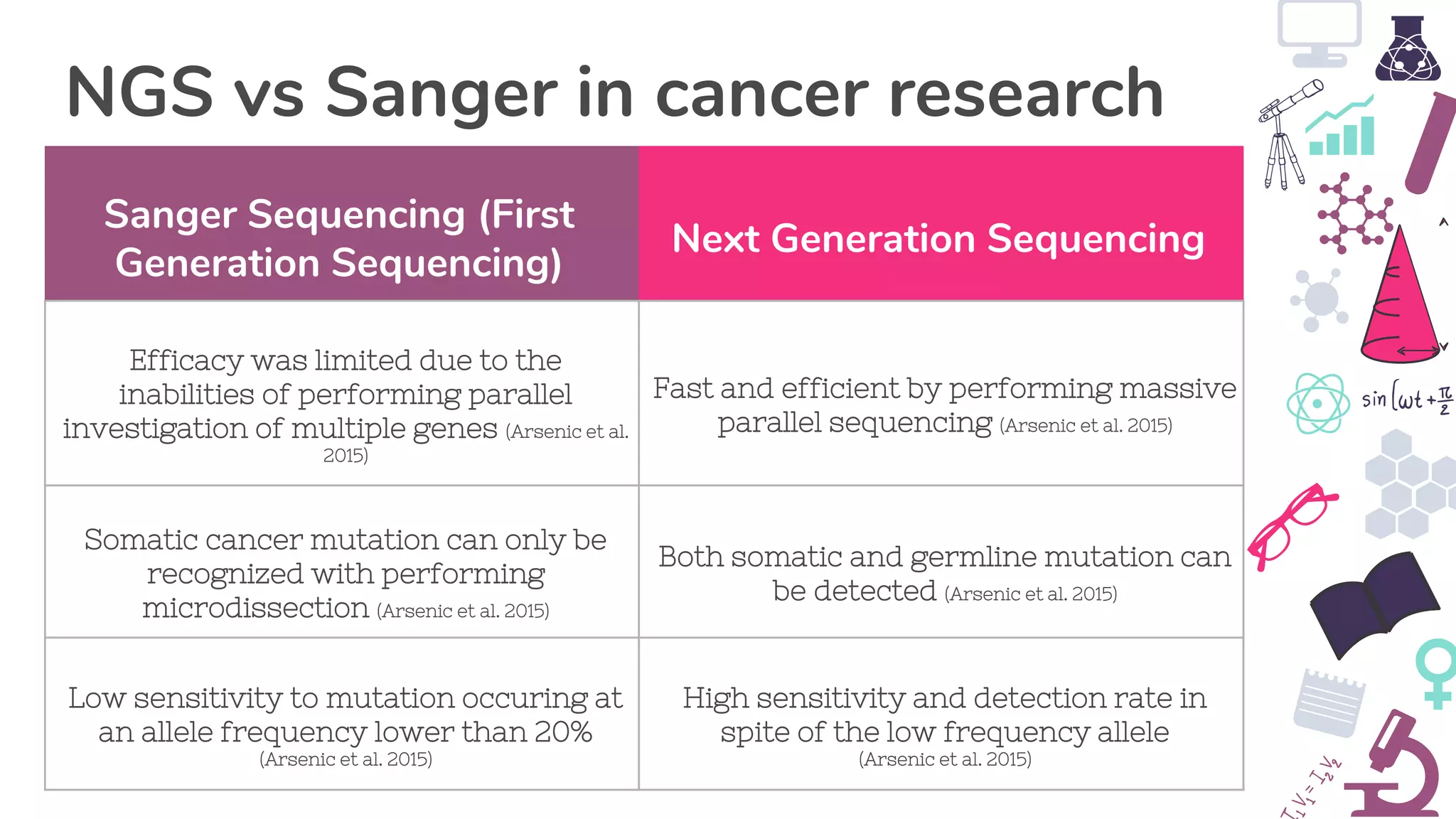
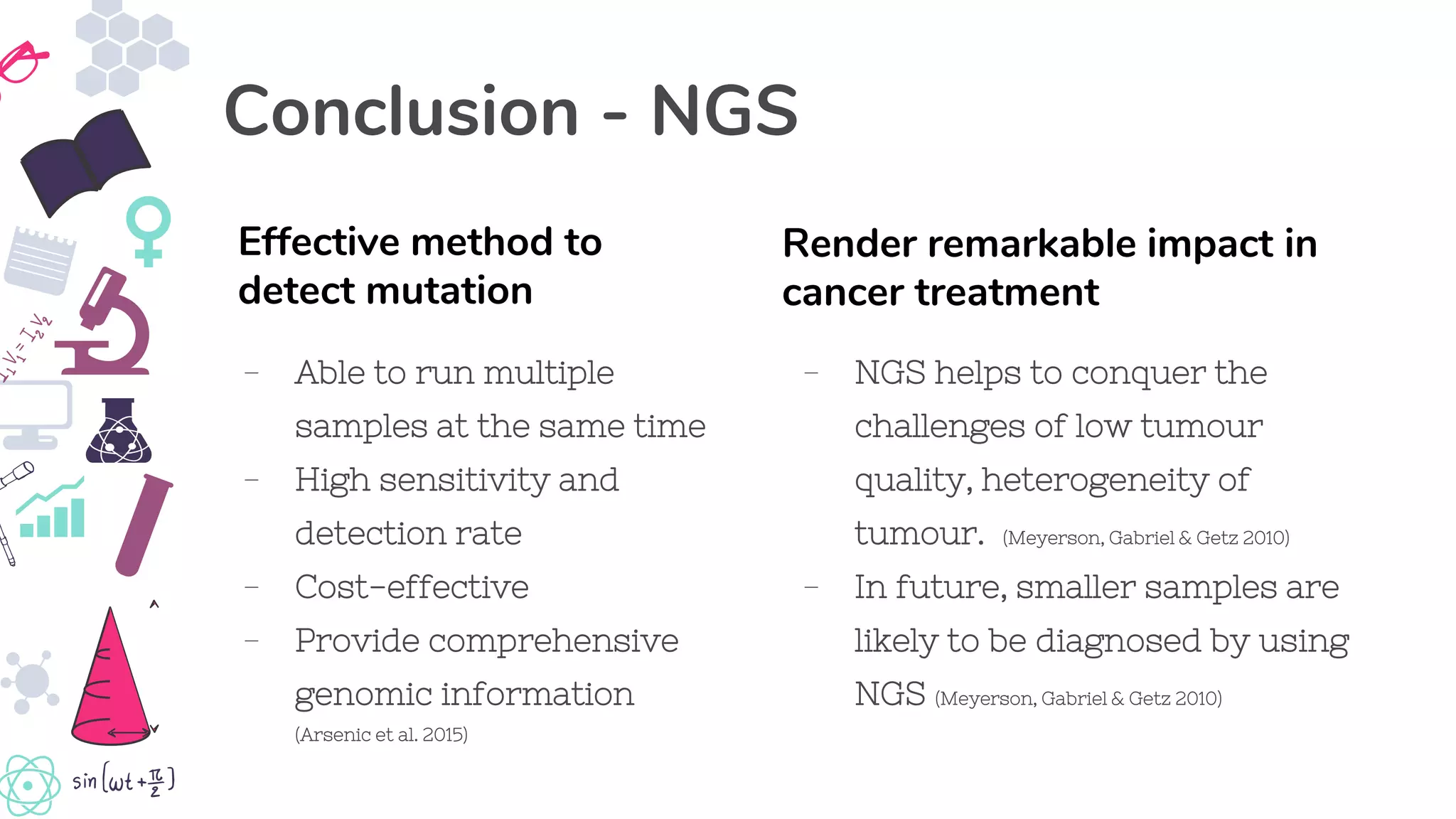
● Arsenic, R, Treue, D, Lehmann, A, Hummel, M, Dietel, M, Denkert, C & Budczies, Jan 2015, ‘Comparison of targeted next-
generation sequencing and Sanger sequencing for the detection of PIK3CA mutations in breast cancer’, BMC clinical
pathology, vol. 15, no. 20, viewed 28 May 2019, <https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4652376/>
● Barba, M., Czosnek, H. and Hadidi, A. 2014. ‘Historical Perspective, Development and Applications of Next-Generation
Sequencing in Plant Virology’. Viruses, [online] 6(1), pp.106-136. Available at:
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3917434/ [Accessed 27 May 2019].
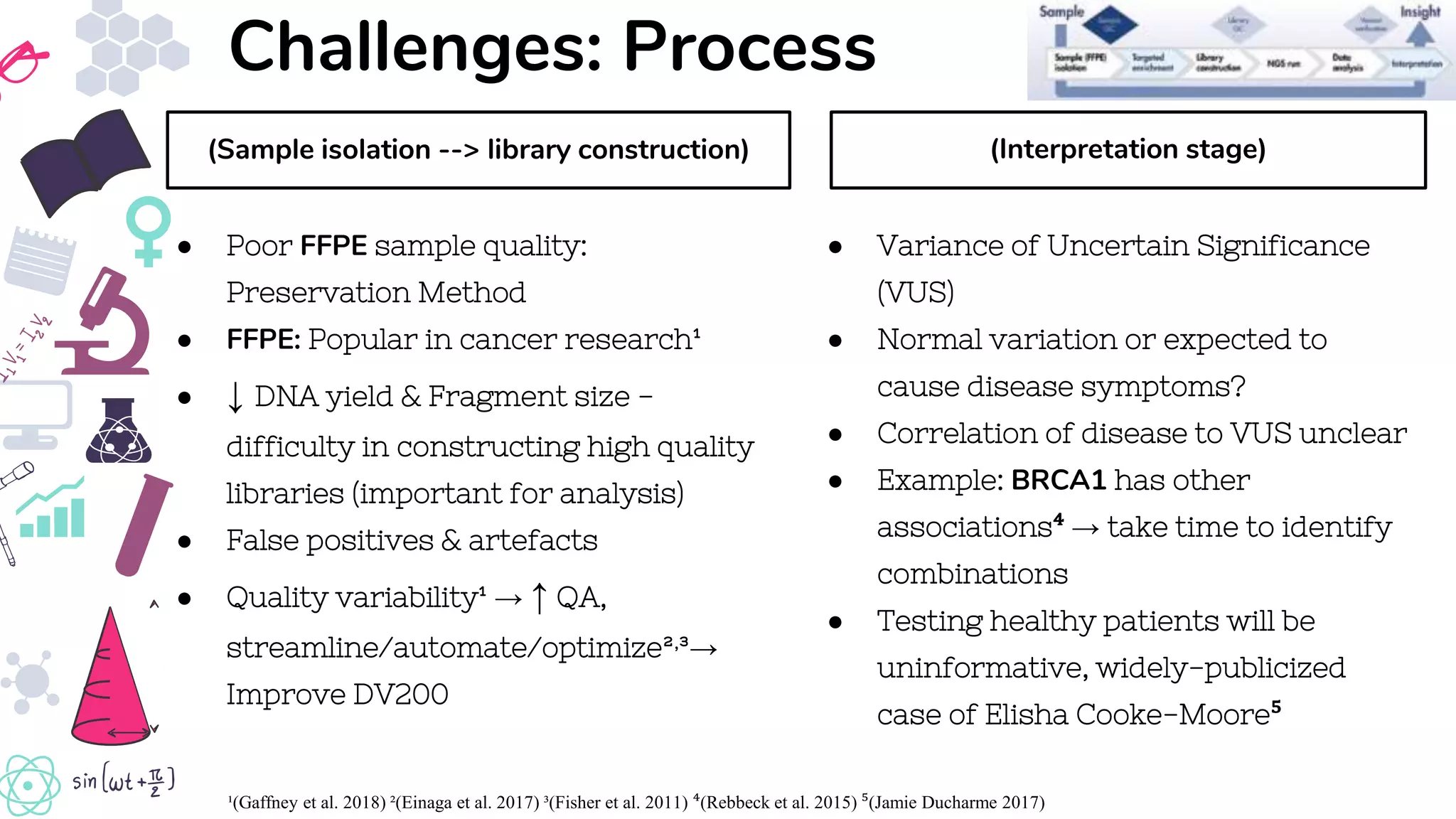
● Einaga, N, Yoshida, A, Noda, H, Suemitsu, M, Nakayama, Y, Sakurada, A, Kawaji, Y, Yamaguchi, H, Sasaki, Y, Tokino, T &
Esumi, M 2017, ‘Assessment of the quality of DNA from various formalin-fixed paraffin-embedded (FFPE) tissues and the
use of this DNA for next-generation sequencing (NGS) with no artifactual mutation’ ed.AWI Lo., PLOS ONE, vol. 12, no. 5,
p.e0176280. Available from: https://dx.plos.org/10.1371/journal.pone.0176280. [27 May 2019].
● Fisher, S, Barry, A, Abreu, J, Minie, B, Nolan, J, Delorey, TM, Young, G, Fennell, TJ, Allen, A, Ambrogio, L, Berlin, AM,
Blumenstiel, B, Cibulskis, K, Friedrich, D, Johnson, R, Juhn, F, Reilly, B, Shammas, R, Stalker, J, Sykes, SM, Thompson, J,
Walsh, J, Zimmer, A, Zwirko, Z, Gabriel, S, Nicol, R & Nusbaum, C 2011, ‘A scalable, fully automated process for construction
of sequence-ready human exome targeted capture libraries’., Genome Biology, vol. 12, no. 1, p.R1. Available from:
http://genomebiology.biomedcentral.com/articles/10.1186/gb-2011-12-1-r1. [28 May 2019].
● Gaffney, E, Riegman, P, Grizzle, W & Watson, P 2018, ‘Factors that drive the increasing use of FFPE tissue in basic and
translational cancer research’., Biotechnic & Histochemistry, vol. 93, no. 5, pp.373–386. Available from:
https://www.tandfonline.com/doi/full/10.1080/10520295.2018.1446101. [27 May 2019].
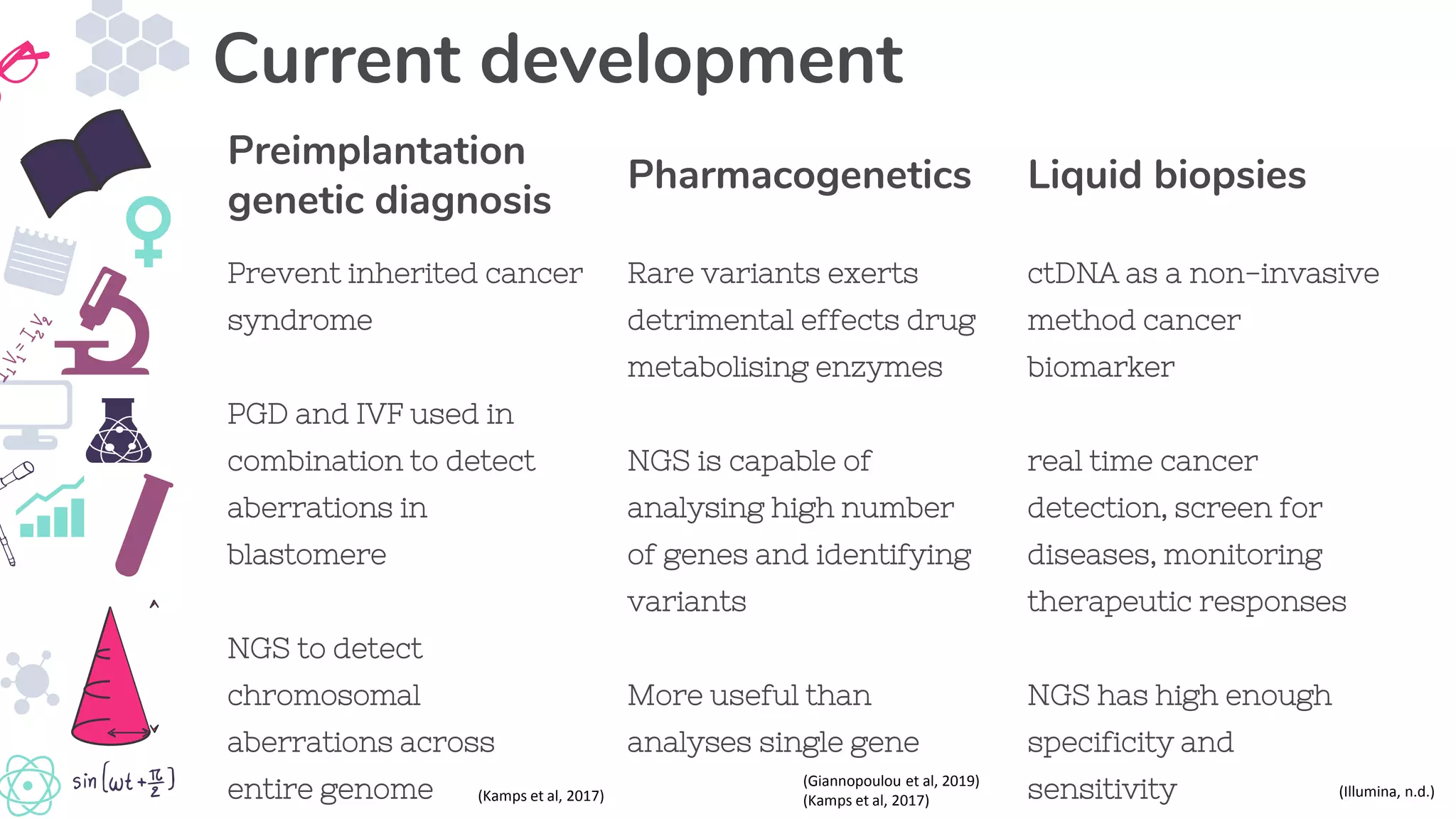
● Giannapoulou, E, Katsila, T, Mitropoulou, C, Tsermpini, EE & Patrinos, GP 2019, ‘Integrating next generation sequencing in
the clinical pharmacogenomins workflow’, Frontiers in Pharmacology, viewed 27 May 2019,
<https://doi.org/10.3389/fphar.2019.00384>
● Heather, J. and Chain, B. 2016. ‘The sequence of sequencers: The history of sequencing DNA’. Genomics, [online] 107(1),
pp.1-8. Available at: https://www.sciencedirect.com/science/article/pii/S0888754315300410 [Accessed 27 May 2019].
● Illumina, n.d., An introduction to next generation sequencing for oncologist, viewed 27 May 2019,
<https://www.illumina.com/content/dam/illumina-marketing/documents/products/other/primer-ngs-oncology.pdf>
References](https://image.slidesharecdn.com/gtcsct60103group5assignment-190528010336/75/Next-Generation-Sequencing-12-2048.jpg)
![● Jamie Ducharme 2017, ‘Woman Sues After Unnecessary Mastectomy and Hysterectomy | Time’ Available from:
http://time.com/4994961/breast-cancer-uterus-surgery/. [27 May 2019].
● Kamps, R., Brandão, R., Bosch, B., Paulussen, A., Xanthoulea, S., Blok, M. and Romano, A. 2017. ‘Next-Generation
Sequencing in Oncology: Genetic Diagnosis, Risk Prediction and Cancer Classification. International Journal of Molecular
Sciences’, [online] 18(2), p.308. Available at: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5343844/ [Accessed 27 May
2019].
● Kulski, J. 2016. ‘Next-Generation Sequencing — An Overview of the History, Tools, and “Omic” Applications. Next
Generation Sequencing - Advances, Applications and Challenges’. [online] Available at:
https://www.intechopen.com/books/next-generation-sequencing-advances-applications-and-challenges/next-generation-
sequencing-an-overview-of-the-history-tools-and-omic-applications [Accessed 27 May 2019].
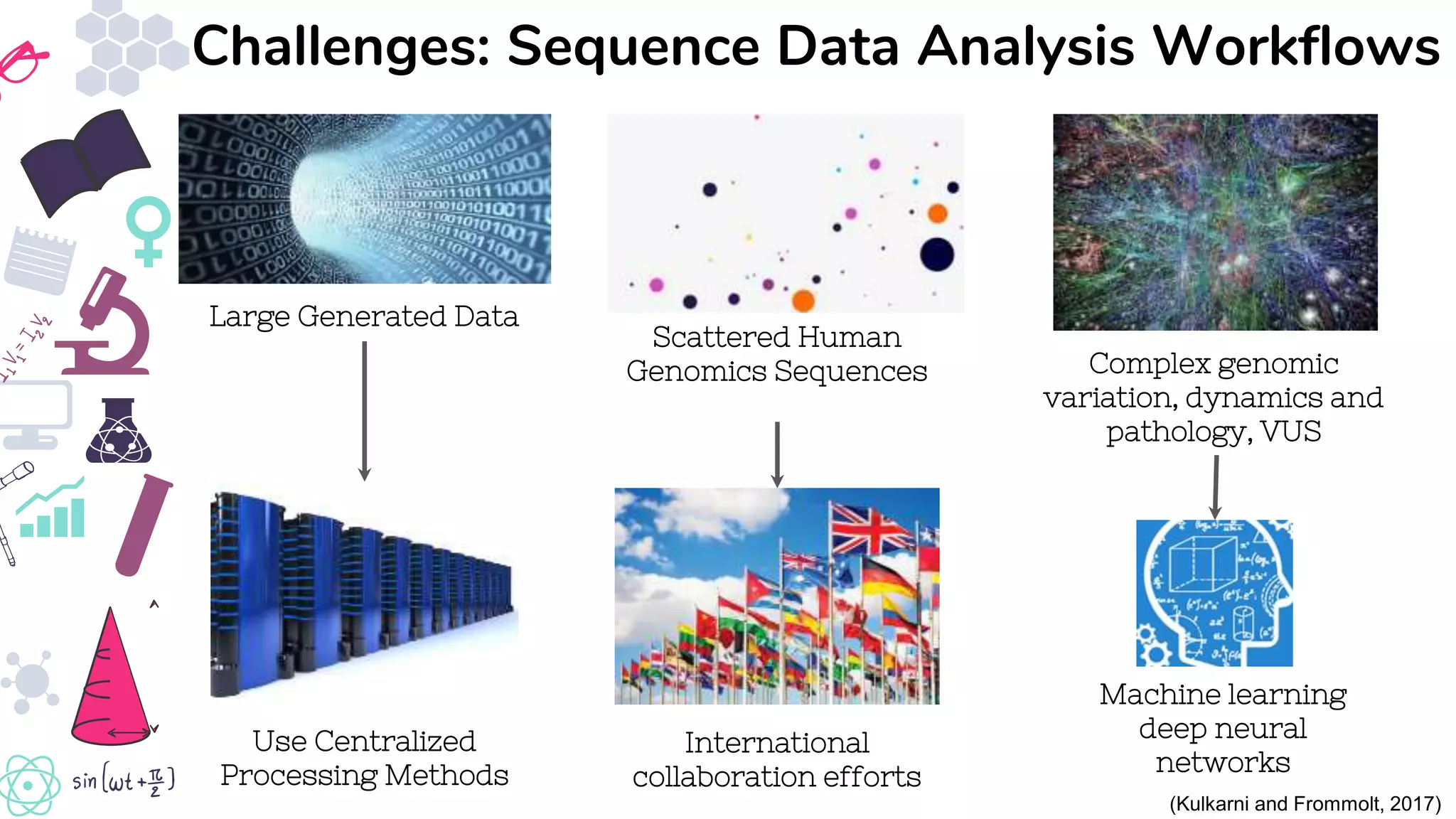
● Kulkarni, P. and Frommolt, P. (2017) ‘Challenges in the Setup of Large-scale Next-Generation Sequencing Analysis
Workflows.’, Computational and structural biotechnology journal. Research Network of Computational and Structural
Biotechnology, 15, pp. 471–477. doi: 10.1016/j.csbj.2017.10.001.
● Nagahashi, M, Shimada, Y, Ichikawa, H, Nakagawa, S, Sato, N, Kaneko, K, Homma, K, Kawasaki, T, Kodama, K, Lyle, S,
Takabe, K Wakai, T 2017, ‘Formalin Fixed paraffin-embedded sample conditions for deep next generation sequencing’,
Journal of Surgical Research, vol, 220, no. 2017, pp. 125-132
● New England BioLabs, n.d., FFPE DNA, viewed 27 May 2019, <https://www.neb.com/products/sample-preparation-for-
next-generation-sequencing/ffpe-dna/ffpe-dna>
● Ozsolak, F. 2012. ‘Third-generation sequencing techniques and applications to drug discovery. Expert Opinion on Drug
Discovery’, [online] 7(3), pp.231-243. Available at:
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3319653/pdf/nihms352467.pdf [Accessed 27 May 2019].
● Płoski, R. 2016. ‘Next Generation Sequencing—General Information about the Technology, Possibilities, and Limitations.
Clinical Applications for Next-Generation Sequencing’ [online] pp.1-18. Available at:
https://www.sciencedirect.com/science/article/pii/B9780128017395000015 [Accessed 27 May 2019].
References](https://image.slidesharecdn.com/gtcsct60103group5assignment-190528010336/75/Next-Generation-Sequencing-13-2048.jpg)
![References
● Rebbeck, TR, Mitra, N, Wan, F, Sinilnikova, OM, Healey, S, McGuffog, L, Mazoyer, S, Chenevix-Trench, G, Easton, DF, Antoniou, AC, Nathanson, KL, Laitman, Y,
Kushnir, A, Paluch-Shimon, S, Berger, R, Zidan, J, Friedman, E, Ehrencrona, H, Stenmark-Askmalm, M, Einbeigi, Z, Loman, N, Harbst, K, Rantala, J, Melin, B, Huo, D,
Olopade, OI, Seldon, J, Ganz, PA, Nussbaum, RL, Chan, SB, Odunsi, K, Gayther, SA, Domchek, SM, Arun, BK, Lu, KH, Mitchell, G, Karlan, BY, Walsh, C, Lester, J,
Godwin, AK, Pathak, H, Ross, E, Daly, MB, Whittemore, AS, John, EM, Miron, A, Terry, MB, Chung, WK, Goldgar, DE, Buys, SS, Janavicius, R, Tihomirova, L, Tung, N,
Dorfling, CM, van Rensburg, EJ, Steele, L, Neuhausen, SL, Ding, YC, Ejlertsen, B, Gerdes, A-M, Hansen, T v. O, Ramón y Cajal, T, Osorio, A, Benitez, J, Godino, J,
Tejada, M-I, Duran, M, Weitzel, JN, Bobolis, KA, Sand, SR, Fontaine, A, Savarese, A, Pasini, B, Peissel, B, Bonanni, B, Zaffaroni, D, Vignolo-Lutati, F, Scuvera, G,
Giannini, G, Bernard, L, Genuardi, M, Radice, P, Dolcetti, R, Manoukian, S, Pensotti, V, Gismondi, V, Yannoukakos, D, Fostira, F, Garber, J, Torres, D, Rashid, MU,
Hamann, U, Peock, S, Frost, D, Platte, R, Evans, DG, Eeles, R, Davidson, R, Eccles, D, Cole, T, Cook, J, Brewer, C, Hodgson, S, Morrison, PJ, Walker, L, Porteous, ME,
Kennedy, MJ, Izatt, L, Adlard, J, Donaldson, A, Ellis, S, Sharma, P, Schmutzler, RK, Wappenschmidt, B, Becker, A, Rhiem, K, Hahnen, E, Engel, C, Meindl, A, Engert, S,
Ditsch, N, Arnold, N, Plendl, HJ, Mundhenke, C, Niederacher, D, Fleisch, M, Sutter, C, Bartram, CR, Dikow, N, Wang-Gohrke, S, Gadzicki, D, Steinemann, D, Kast, K,
Beer, M, Varon-Mateeva, R, Gehrig, A, Weber, BH, Stoppa-Lyonnet, D, Sinilnikova, OM, Mazoyer, S, Houdayer, C, Belotti, M, Gauthier-Villars, M, Damiola, F, Boutry-
Kryza, N, Lasset, C, Sobol, H, Peyrat, J-P, Muller, D, Fricker, J-P, Collonge-Rame, M-A, Mortemousque, I, Nogues, C, Rouleau, E, Isaacs, C, De Paepe, A, Poppe, B,
Claes, K, De Leeneer, K, Piedmonte, M, Rodriguez, G, Wakely, K, Boggess, J, Blank, S V., Basil, J, Azodi, M, Phillips, K-A, Caldes, T, de la Hoya, M, Romero, A,
Nevanlinna, H, Aittomäki, K, van der Hout, AH, Hogervorst, FBL, Verhoef, S, Collée, JM, Seynaeve, C, Oosterwijk, JC, Gille, JJP, Wijnen, JT, Garcia, EBG, Kets, CM,
Ausems, MGEM, Aalfs, CM, Devilee, P, Mensenkamp, AR, Kwong, A, Olah, E, Papp, J, Diez, O, Lazaro, C, Darder, E, Blanco, I, Salinas, M, Jakubowska, A, Lubinski, J,
Gronwald, J, Jaworska-Bieniek, K, Durda, K, Sukiennicki, G, Huzarski, T, Byrski, T, Cybulski, C, Toloczko-Grabarek, A, Zlowocka-Perlowska, E, Menkiszak, J, Arason, A,
Barkardottir, RB, Simard, J, Laframboise, R, Montagna, M, Agata, S, Alducci, E, Peixoto, A, Teixeira, MR, Spurdle, AB, Lee, MH, Park, SK, Kim, S-W, Friebel, TM,
Couch, FJ, Lindor, NM, Pankratz, VS, Guidugli, L, Wang, X, Tischkowitz, M, Foretova, L, Vijai, J, Offit, K, Robson, M, Rau-Murthy, R, Kauff, N, Fink-Retter, A, Singer,
CF, Rappaport, C, Gschwantler-Kaulich, D, Pfeiler, G, Tea, M-K, Berger, A, Greene, MH, Mai, PL, Imyanitov, EN, Toland, AE, Senter, L, Bojesen, A, Pedersen, IS,
Skytte, A-B, Sunde, L, Thomassen, M, Moeller, ST, Kruse, TA, Jensen, UB, Caligo, MA, Aretini, P, Teo, S-H, Selkirk, CG, Hulick, PJ, Andrulis, I & Andrulis, I 2015,
‘Association of Type and Location of BRCA1 and BRCA2 Mutations With Risk of Breast and Ovarian Cancer’., JAMA, vol. 313, no. 13, p.1347. Available from:
http://www.ncbi.nlm.nih.gov/pubmed/25849179. [27 May 2019].](https://image.slidesharecdn.com/gtcsct60103group5assignment-190528010336/75/Next-Generation-Sequencing-14-2048.jpg)
![● Serrati, S, Summa, SD, Pilato, B, Patriella, D, Lacalamita, S, Tommasi, S & Pinto, R 2016, ‘Next-generation sequencing:
advances and applications in cancer diagnosis’, OncoTargets and Therapy, no.9, pp. 7355-7365, viewed 28 May 2019,
<https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5144906/>
● Totomoch-Serra, A., Marquez, M. and Cervantes-Barragán, D. 2017. ‘Sanger sequencing as a first-line approach for
molecular diagnosis of Andersen-Tawil syndrome’. F1000Research, [online] 6, p.1016. Available at:
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5635448/pdf/f1000research-6-12541.pdf [Accessed 27 May 2019].
● Meyerson, M, Gabriel, S & Getz, G 2010, ‘Advances in understanding cancer genomes through second-generation
sequencing’, Nature Reviews Genetics, vol.11, no. 10, pp. 685-696, viewed 28 May 2019,
<http://ccsb.stanford.edu/content/dam/sm/ccsb/documents/education/cbio243course/2010_Meyerson_CancerNGS_NRG.pdf
>
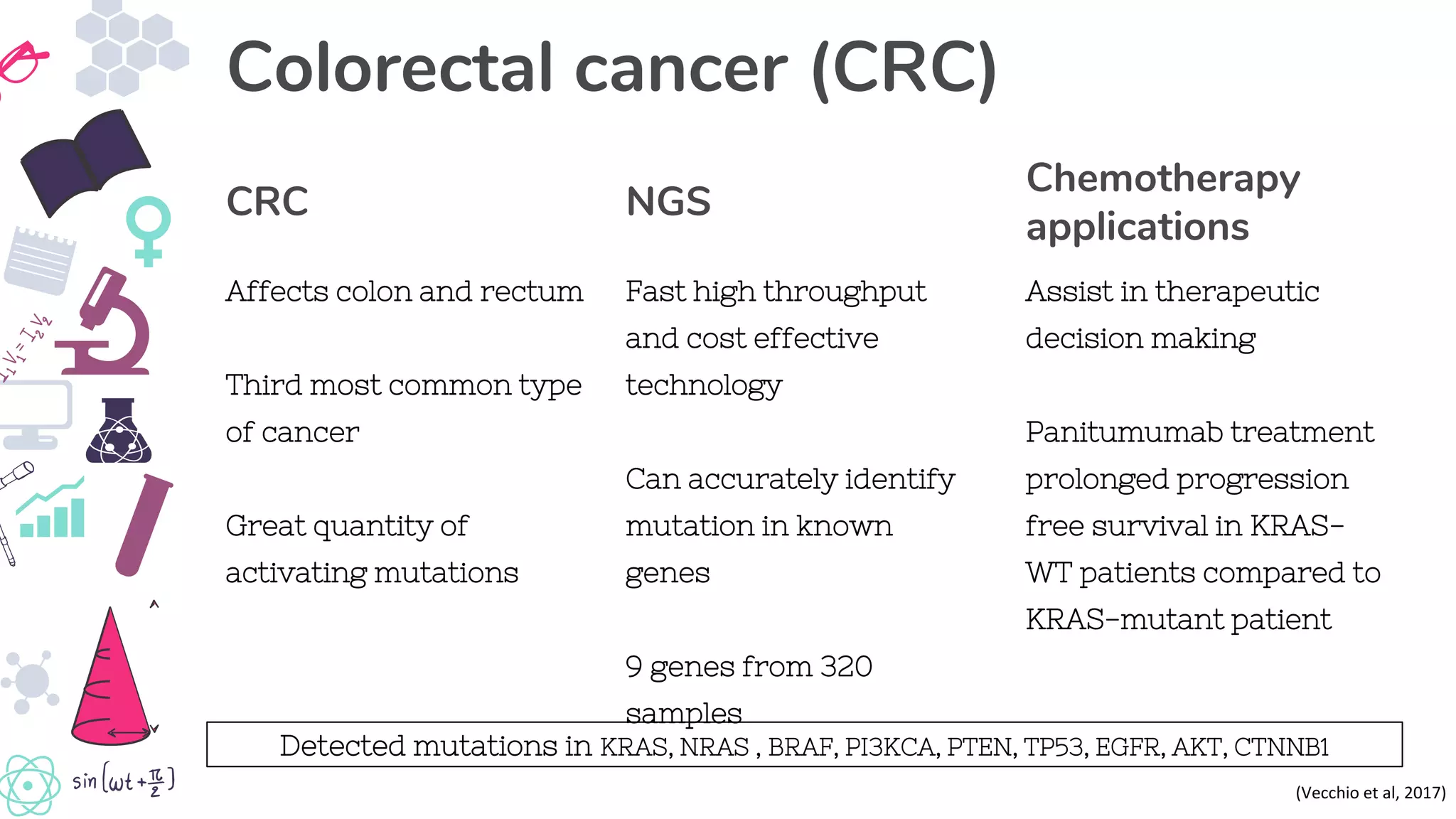
● Vecchio, FD, Mastroiaco, V, Marco, AD, Compagnoni, C, Capece, D, Zazzeroni, F, Capalbo, C, Alesse, E & Tessitore, A 2017,
‘Next generation sequencing: recent applications to the analysis of colorectal cancer’, Journal or Translational Medicine, vol.
15, no. 246. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5723063/ [28 May 2019]
References](https://image.slidesharecdn.com/gtcsct60103group5assignment-190528010336/75/Next-Generation-Sequencing-15-2048.jpg)