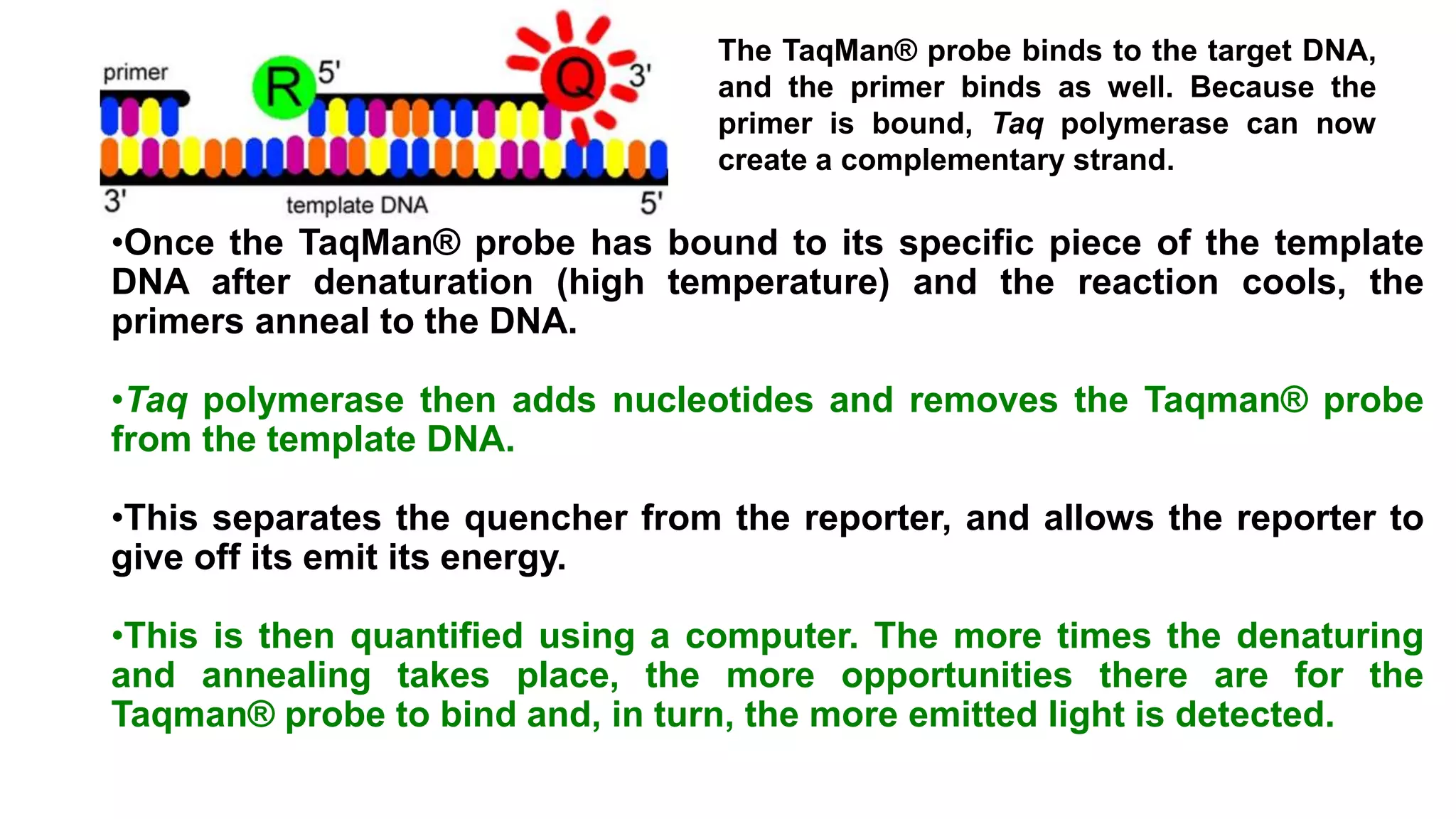
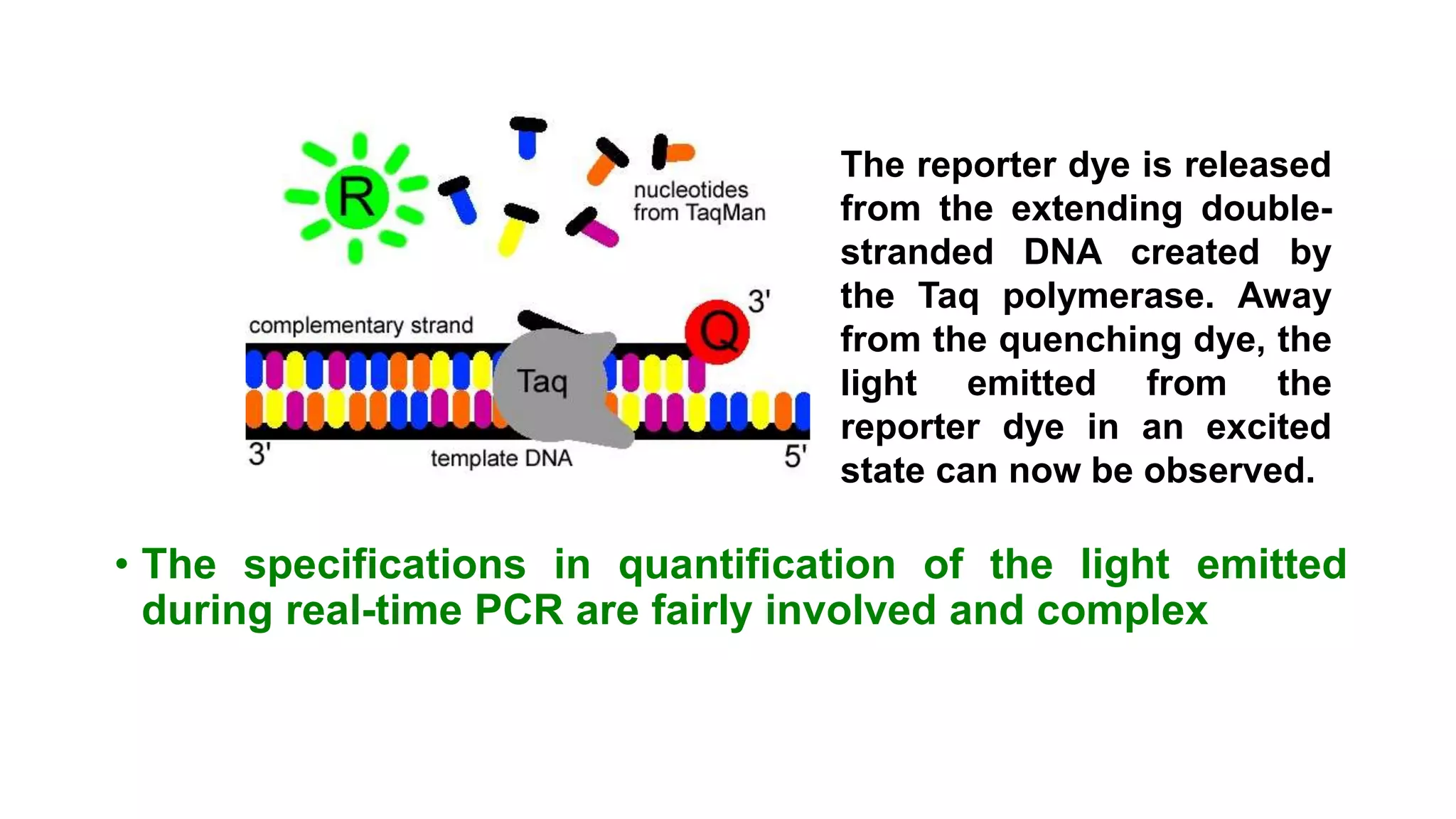
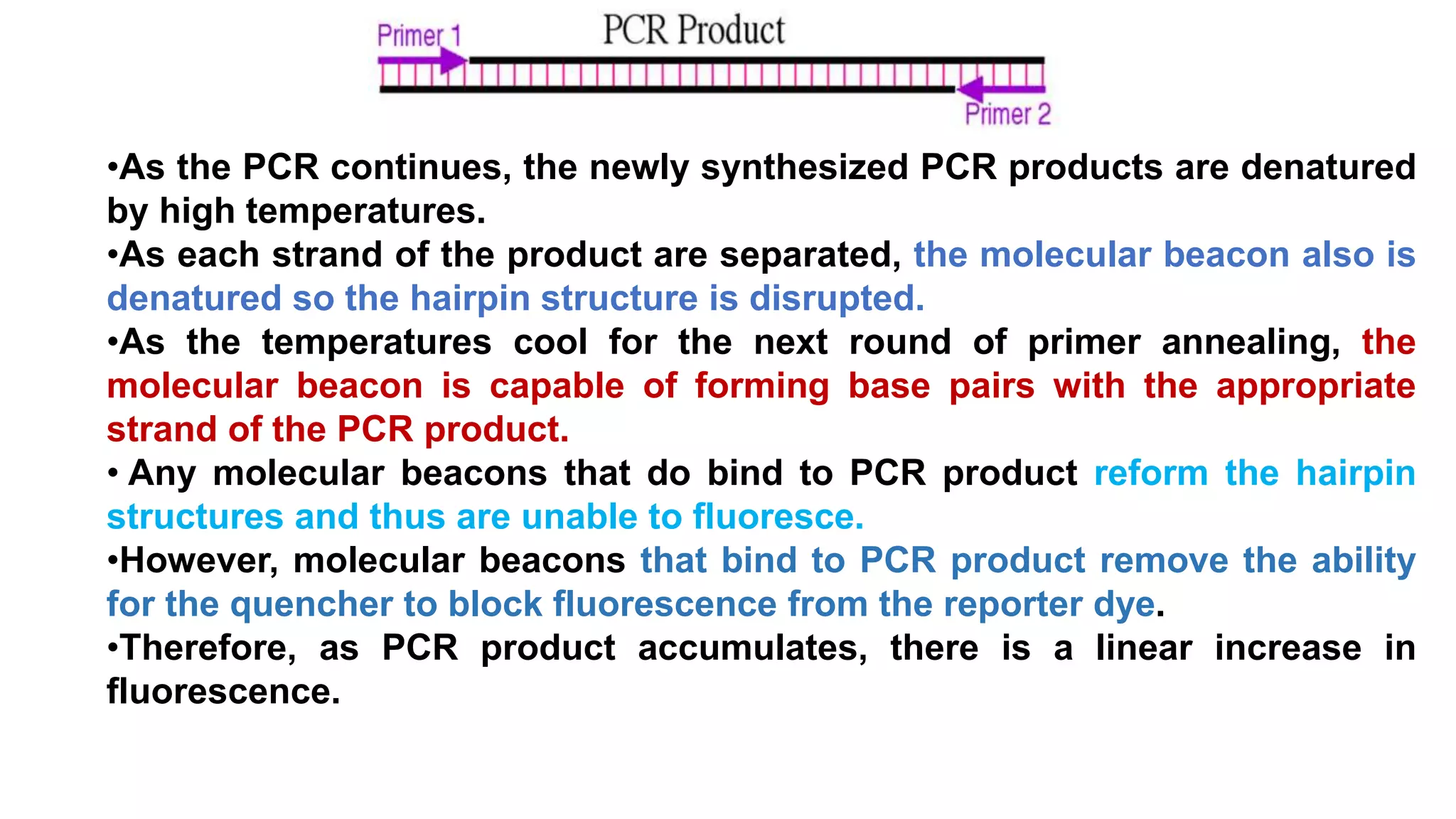
Real-time PCR is an advanced method allowing the visualization and quantification of DNA during amplification, offering advantages over standard PCR, such as higher sensitivity and the ability to detect reactions in progress. It utilizes fluorescent probes to measure PCR products in real-time, with techniques like TaqMan and molecular beacons enhancing specific detection. The method is applicable in various fields including gene expression quantification and pathogen detection, despite its higher technical demands and operational costs.





![• The probe consists of two types of fluorophores, which are the fluorescent
parts of reporter proteins [Green Fluorescent Protein (GFP) has an often-
used fluorophore].
• While the probe is attached or unattached to the template DNA and before
the polymerase acts, the quencher (Q) fluorophore (usually a long-
wavelength colored dye, such as red) reduces the fluorescence from the
reporter (R) fluorophore (usually a short-wavelength colored dye, such as
green).
• It does this by the use of Fluorescence (or Förster) Resonance Energy
Transfer (FRET), which is the inhibition of one dye caused by another
without emission of a proton.
• The reporter dye is found on the 5’ end of the probe and the quencher at the
3’ end.
The Taqman probe. The red circle represents the
quenching dye that disrupts the observable signal
from the reporter dye (green circle) when it is within a
short distance](https://image.slidesharecdn.com/realtime-pcr-171125142754/75/Real-Time-PCR-6-2048.jpg)