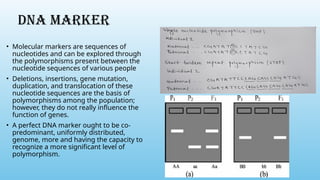
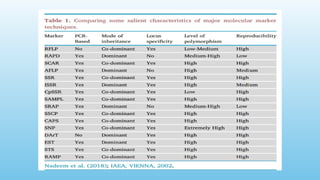
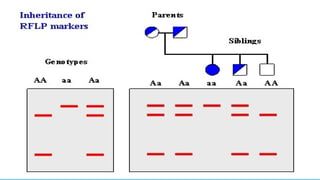
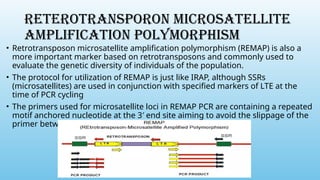
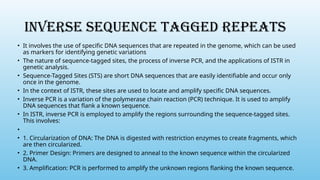
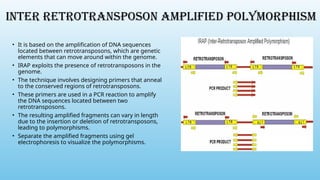
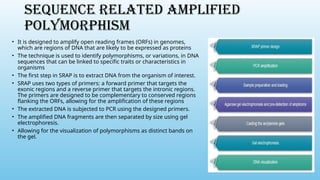
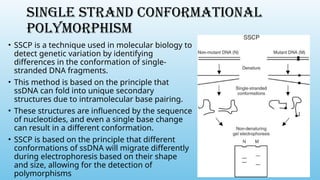
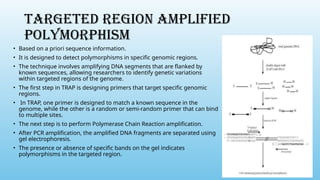
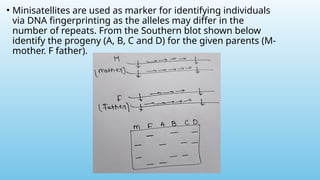
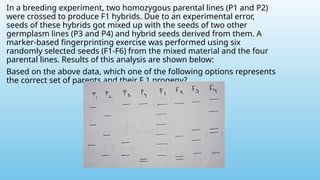
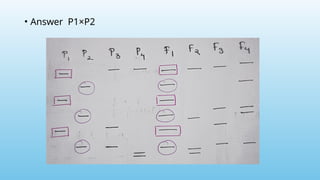
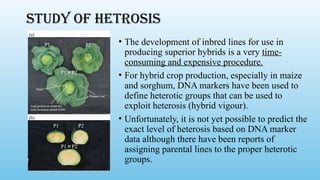
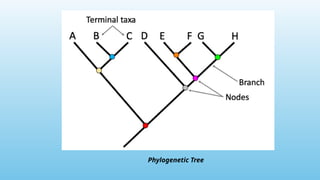
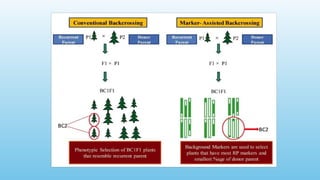
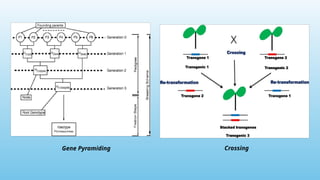
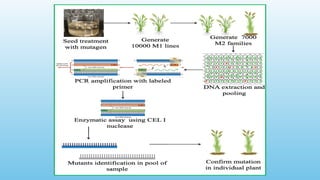
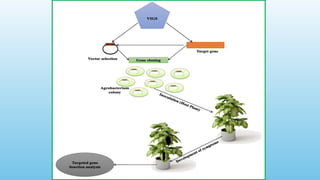
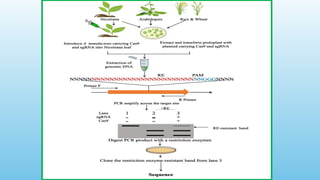
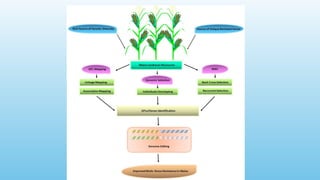
Advanced molecular markers enhance the detection of genetic variations and traits, significantly improving plant breeding techniques like marker-assisted selection (MAS). They offer advantages such as high polymorphism, co-dominance, and efficiency in identifying desired traits earlier in the breeding process. Various techniques, including PCR-based and retrotransposon markers, enable researchers to assess genetic diversity and stability across different species.