- The document describes maximum likelihood estimation (MLE) of species parameters from beetle mass, length, and other character data.
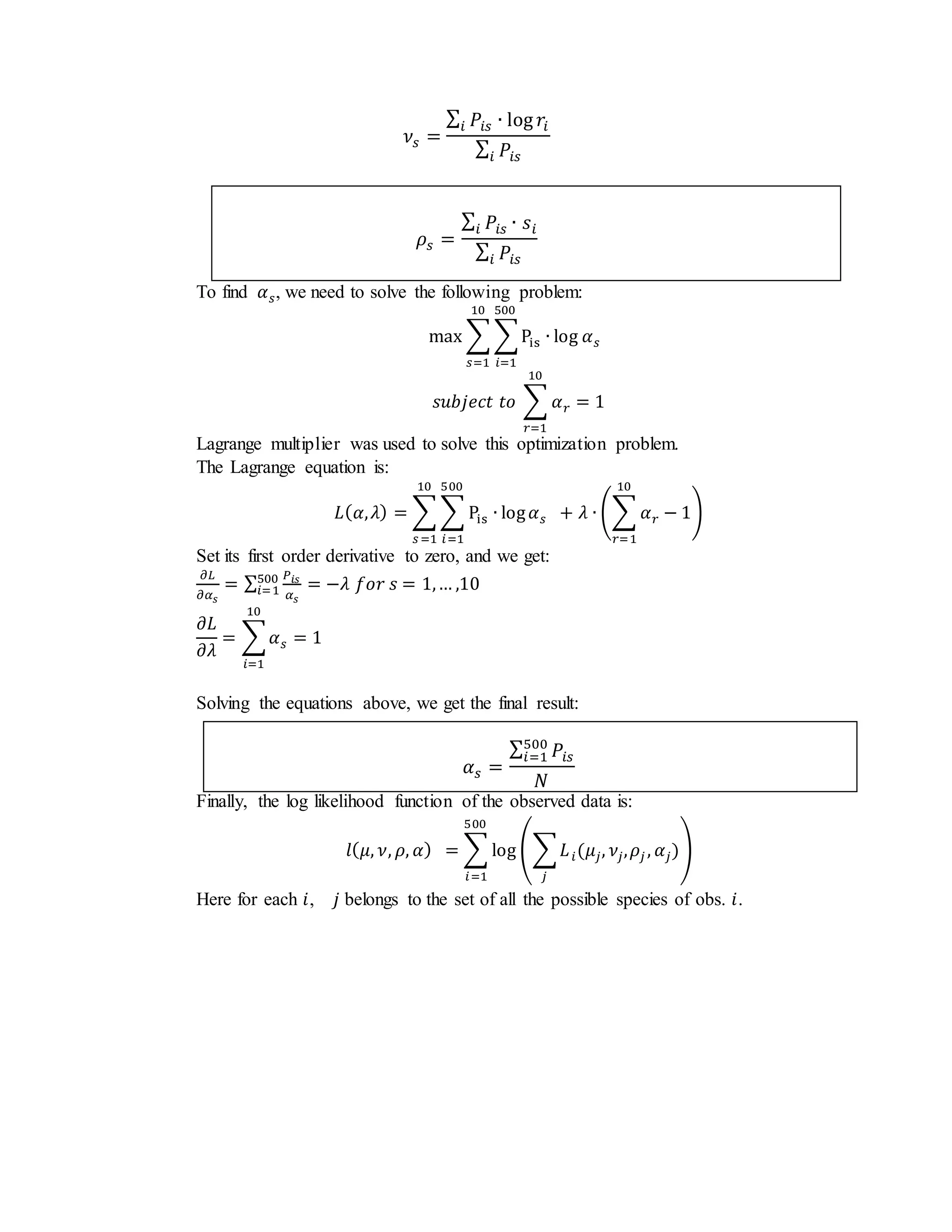
- It derives EM steps to estimate species means (μ, ν), proportions (ρ), and priors (α) in the presence of missing species data.
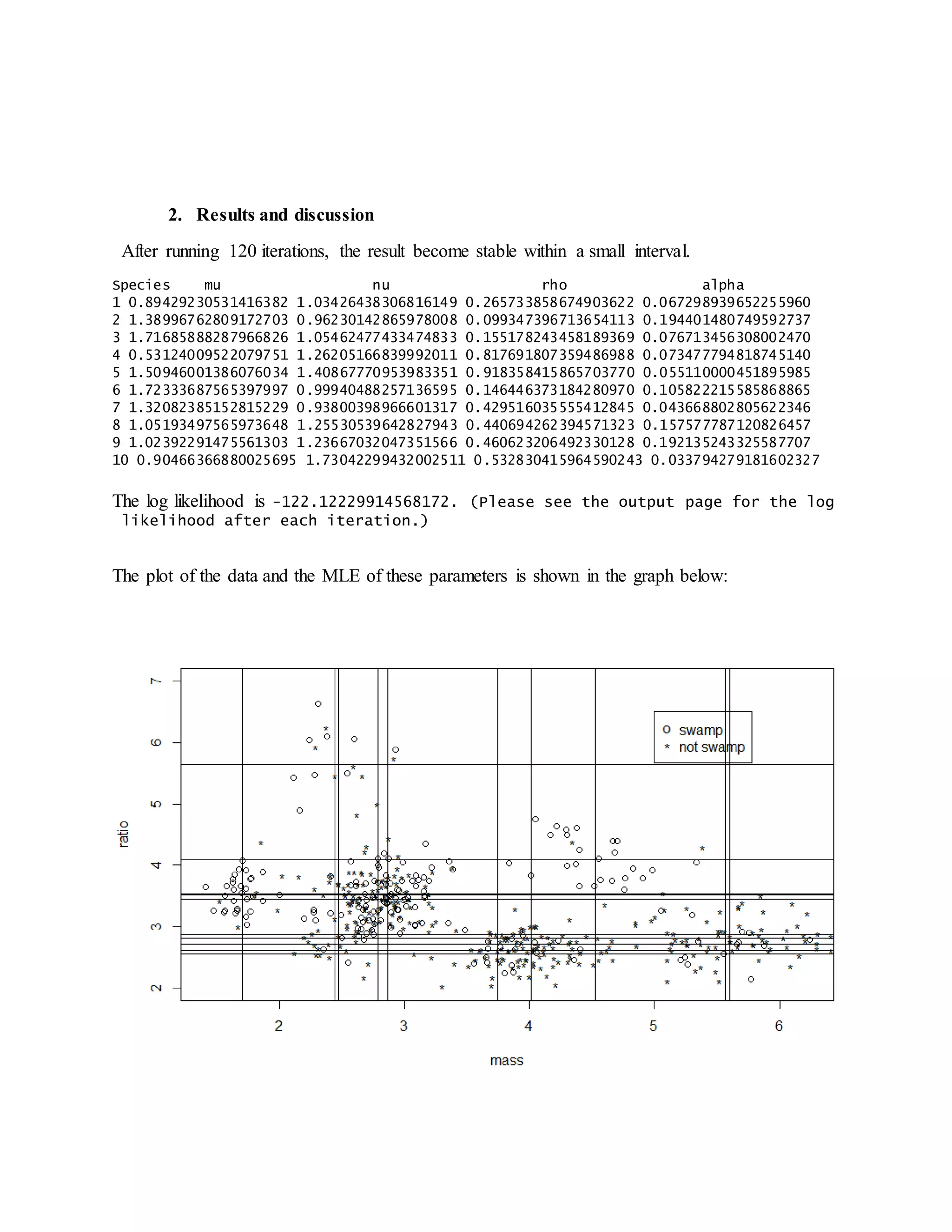
- Running the EM algorithm for 120 iterations estimates the parameters and converges the log likelihood to 14 digits of precision with a convergence rate of approximately 1.
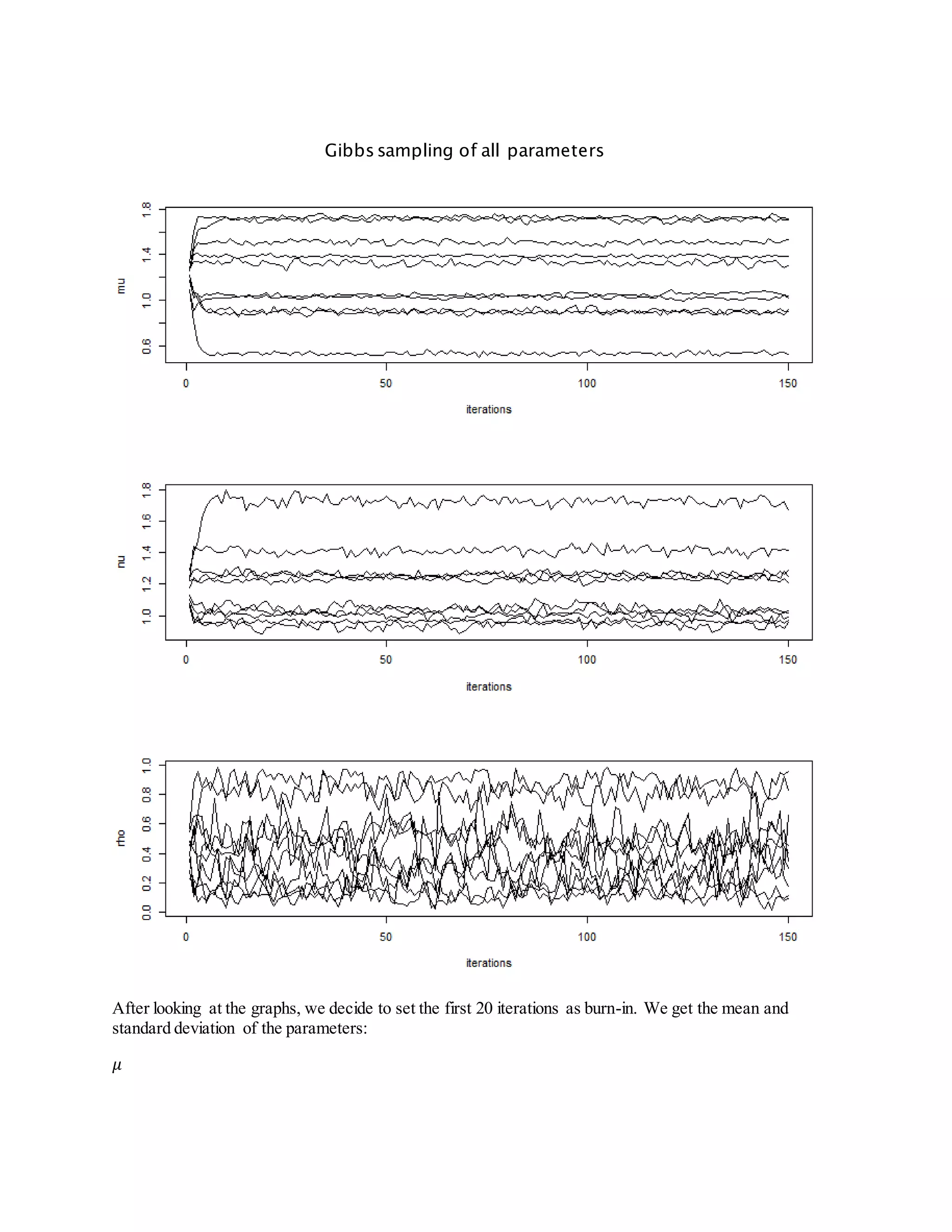
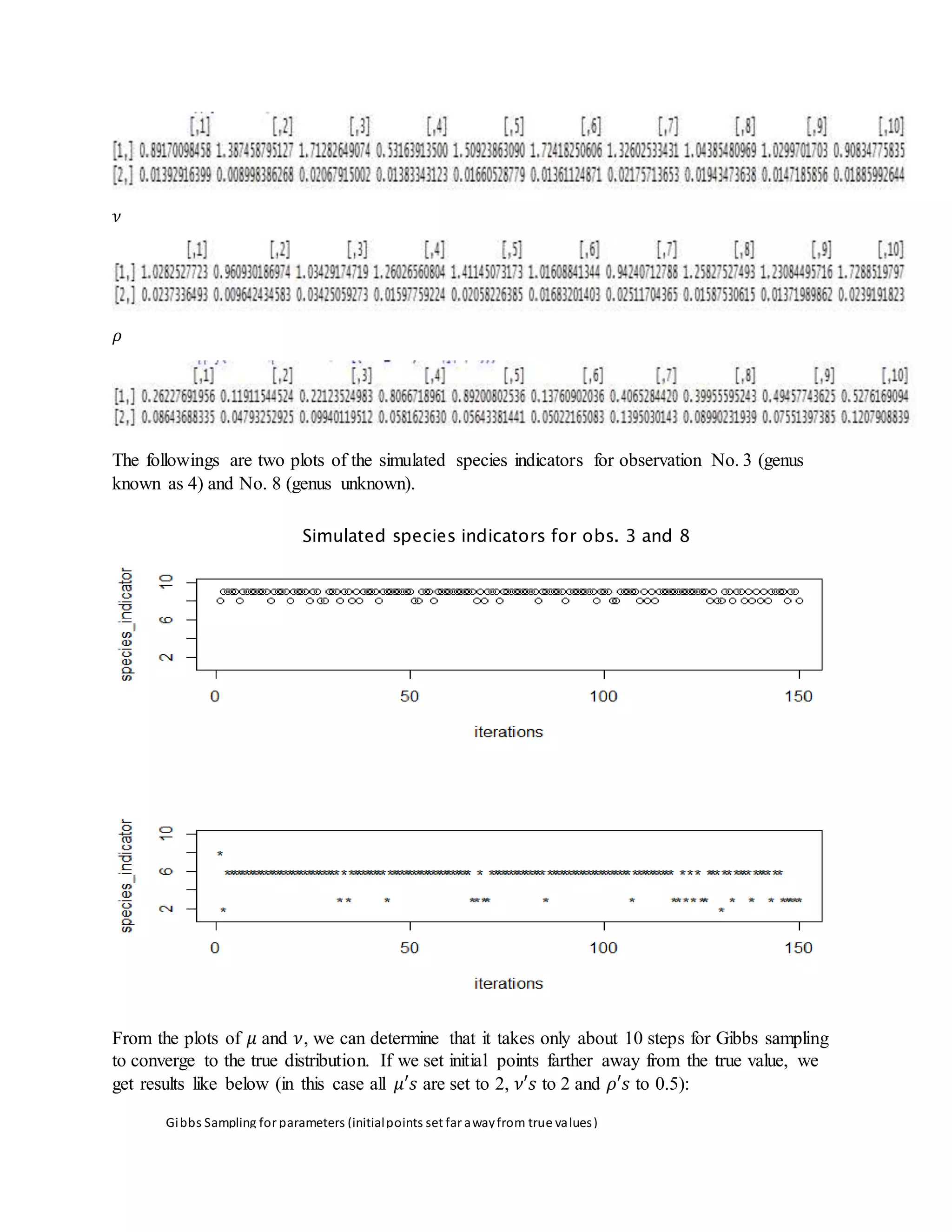
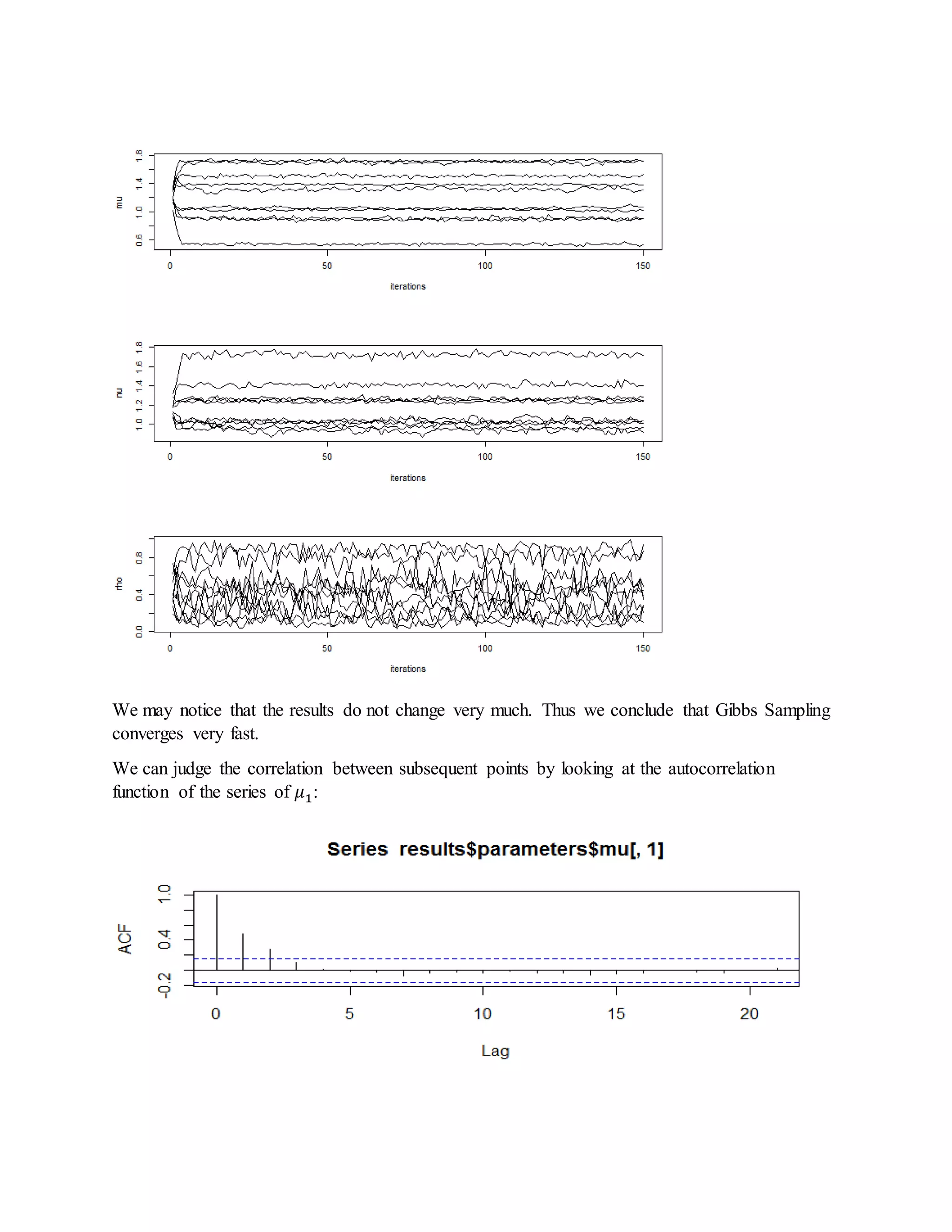
- It also derives steps for Gibbs sampling to estimate the missing species indicators and parameter values based on their posterior distributions.