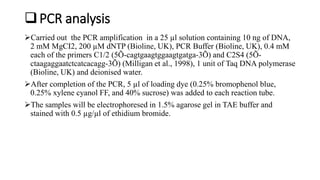
1) Marker assisted selection (MAS) is an indirect selection process where plant breeders select for traits linked to DNA markers rather than the traits themselves.
2) Molecular markers, such as strings of DNA sequences, are linked to desired genes and can be used to identify plants with particular genes through their genotype rather than phenotype.
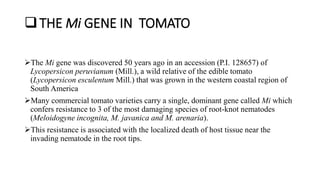
3) For breeding nematode resistance, the Mi gene from a wild tomato species confers resistance to several root-knot nematode species and has been mapped on chromosome 6 along with other linked markers used to select for the gene through MAS.






![DNA isolation
DNA was extraction using the method by Doyle and Doyle (1987)
Freeze fresh leaf tissue in liquid nitrogen and ground using an Eppendorf tube
and glass bar.
Place the homogenate of the leaf tissue in a tube together with 500 μl of DNA
extraction buffer [2% (w/v) CTAB (hexadecyltrimethylammoniumbromide), 1.4
M NaCl, 0.2% (v/v) 2-mercaptoethanol, 20 mM EDTA, and 100 mM Tris-HCl,
pH 8.0], and incubated in a water bath at 600 C for 30 min with occasional
swirling.
After incubation, extract the lysate once with chloroform: phenol (1:1, v/v). Mix
the aqueous phase with a two-thirds volume of cold isopropanol.
Resuspended the precipitated DNA was in TE (10 mM Tris pH 8.0, and 0.1 mM
EDTA)](https://image.slidesharecdn.com/mas-160428095419/85/marker-assisted-selection-in-breeding-for-nematode-resistance-9-320.jpg)