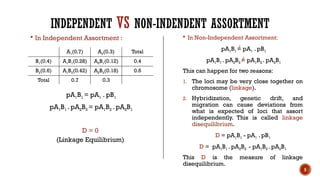
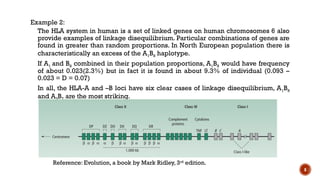
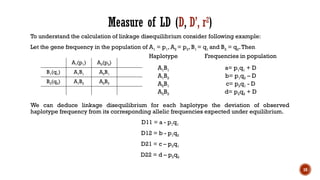
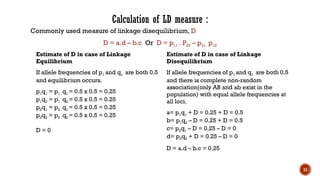
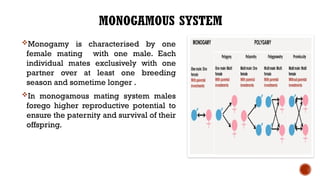
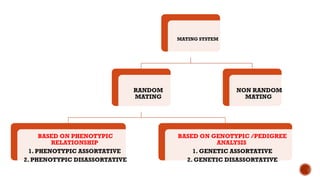
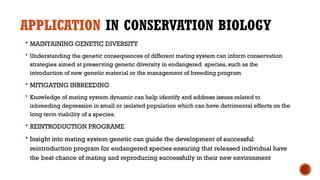
This document provides an overview of linkage disequilibrium (LD) and mating systems, detailing their definitions, significance, and examples. Linkage disequilibrium refers to the non-random association of alleles at different loci, influenced by factors such as selection and recombination rates, while mating systems describe the patterns of mate selection within populations. The document discusses various mating strategies, including monogamy, polygyny, and polyandry, and their evolutionary implications.