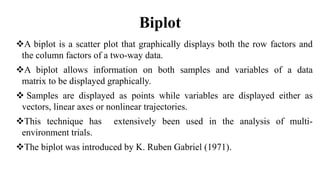
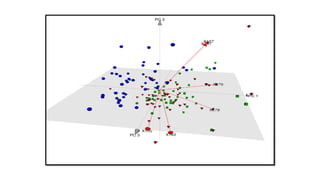
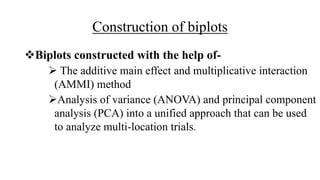
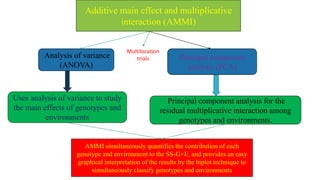
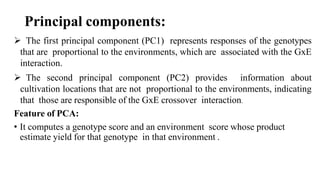
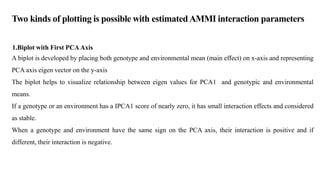
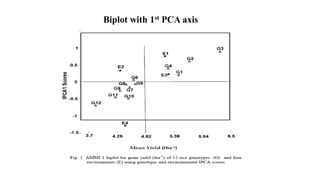
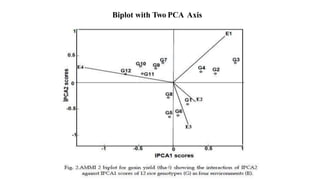
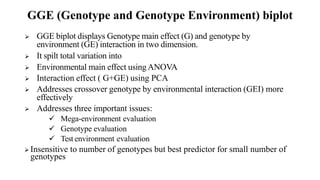
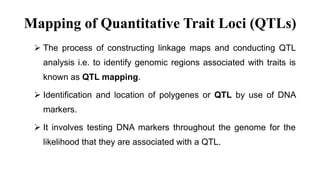
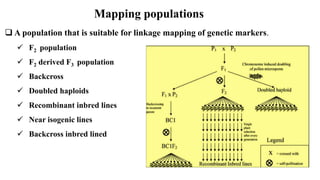
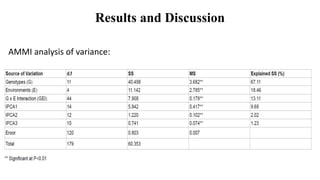
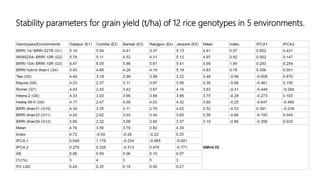
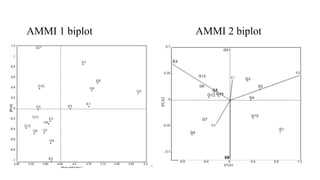
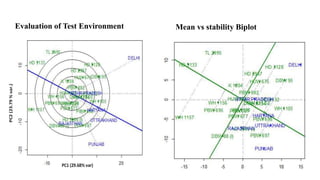
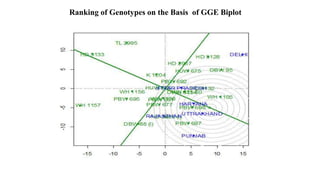
The document discusses using biplots and genotype mapping to analyze multi-environment trials. It describes biplots, how they are constructed using methods like AMMI analysis of variance and principal component analysis. Biplots can show the relationship between genotypes and environments, and identify stable genotypes. The document also discusses genotype by genotype environment (GGE) biplots and their use in identifying mega-environments and ranking genotypes. It provides an example study using these methods to analyze rice hybrids in different locations and identify high yielding stable varieties.