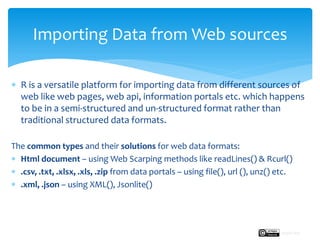
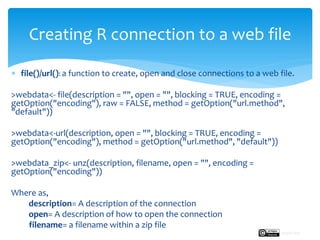
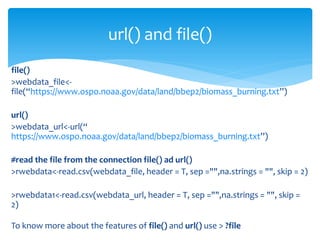
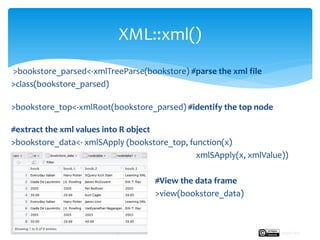
The document provides a comprehensive guide on importing web data into R from various formats including HTML, CSV, XML, and JSON. It describes functions and methods such as readlines(), read.csv(), and jsonlite() for data extraction, parsing, and handling web APIs, alongside web scraping techniques. Additionally, it emphasizes the structure of XML and JSON data formats, further illustrating practical examples for effective data manipulation in R.







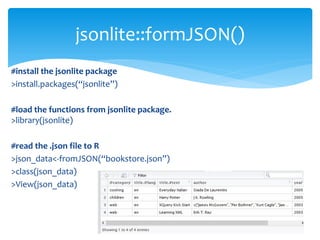



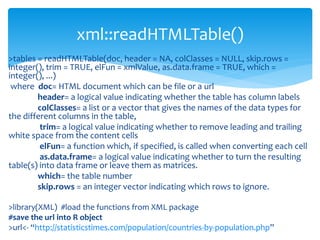
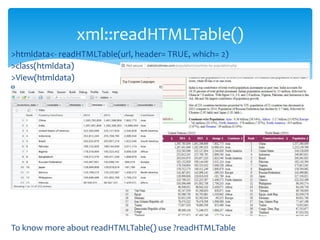
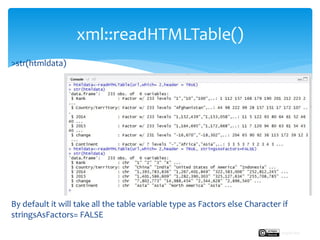

![ For basic web scraping tasks the readLines() and the read.csv functions are
usually sufficient. However this functions allow simple access to webpage
source data on a non-secure servers i.e. http.
For example: web_page <- read.csv("http://www.vulture.com/2018/09/the-best-movies-
of-2018.html")
#grab the required line starting with <em> tag using grep()
>movie_lines <- web_page[grep("<em>", web_page$children)]
#now delete unwanted characters i.e. <em> tag in the lines
>movies2018 <- gsub("<em>", "", author_lines, fixed = TRUE)
#view the best 2018 movie list
>View(movies2018)
readLines() and read.csv()](https://image.slidesharecdn.com/7importwebdata-220113074955/85/Import-web-resources-using-R-Studio-20-320.jpg)

