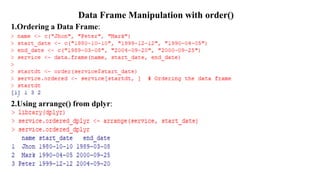
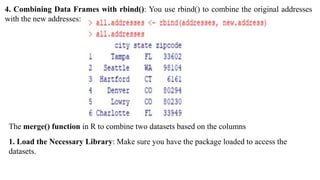
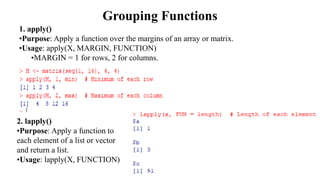
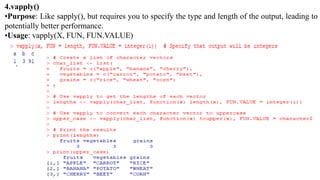
The document provides an overview of data analytics using R, focusing on the handling of datasets and various file types such as CSV, XML, JSON, and Excel. It covers functions for importing and exporting data, manipulating strings, and reshaping data, along with packages for database connectivity and data cleaning. Additionally, it includes examples of using SQL queries and the application of various functions for data processing tasks.

































![Selecting Columns and Rows
1.Selecting Specific Columns, Selecting Specific Rows:
Sorting and Ordering
1.Sorting Vectors:
2.Using order():
x <- c(5, 2, 8, 1, 4)
order_indices <- order(x)
sorted_x <- x[order_indices]
print(sorted_x) # Output: 1 2 4 5 8](https://image.slidesharecdn.com/m3dar-241128130542-cf78bbc6/85/Data-analystics-with-R-module-3-cseds-vtu-37-320.jpg)