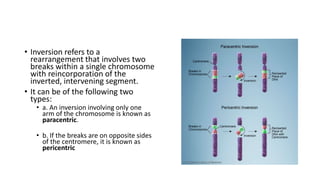
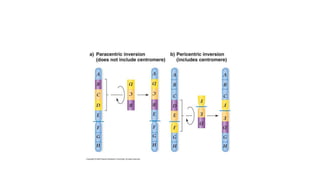
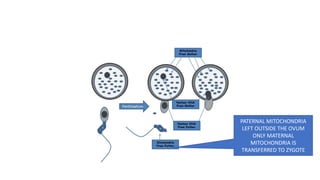
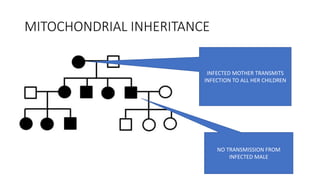
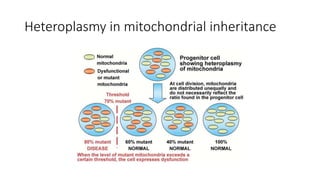
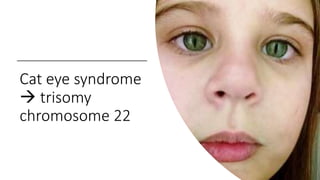
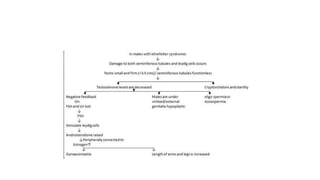
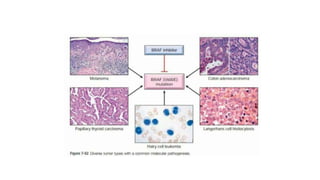
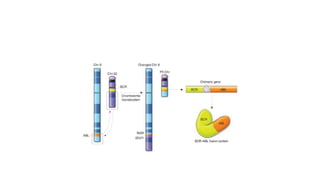
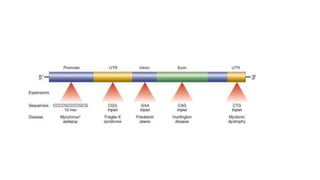
This document discusses various topics in genetics and genomics including:
- Genome and genomics, which refers to the study of the entire genetic content of an individual.
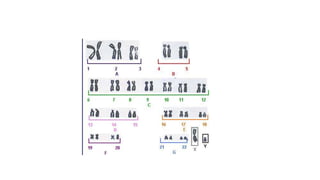
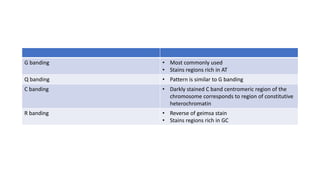
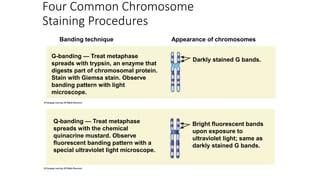
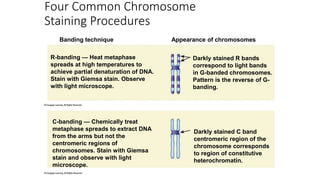
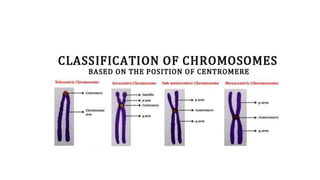
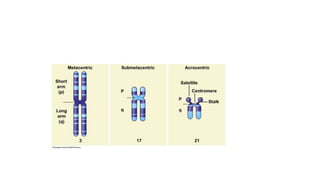
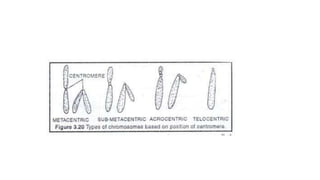
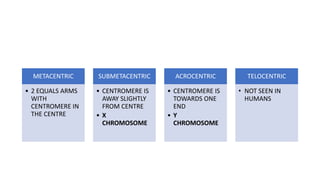
- Karyotyping techniques used to analyze chromosomes such as G-banding and C-banding.
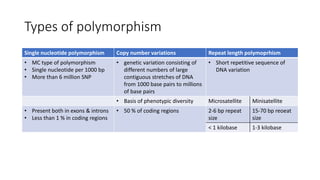
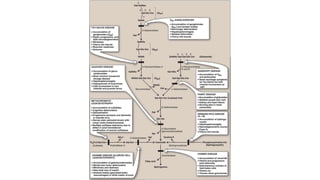
- Types of genetic variation like single nucleotide polymorphisms and copy number variations.
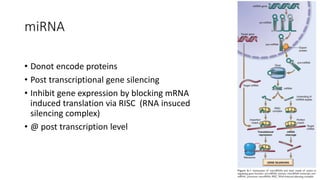
- Non-coding RNAs including microRNAs and long non-coding RNAs that regulate gene expression.
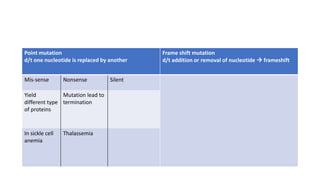
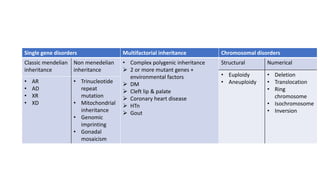
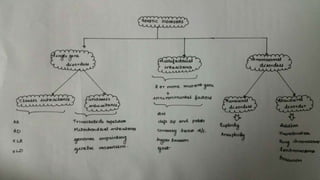
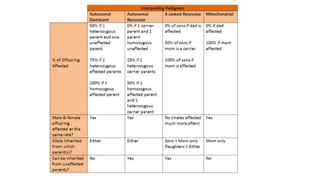
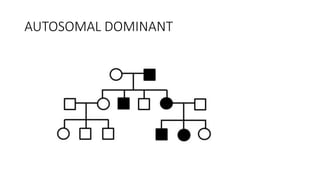
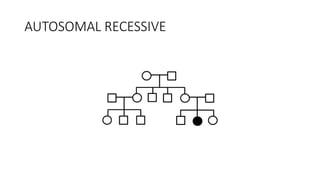
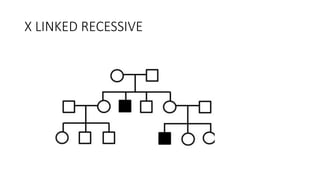
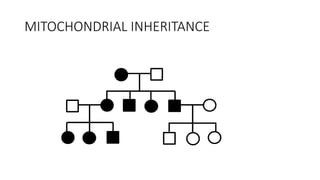
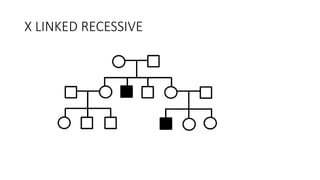
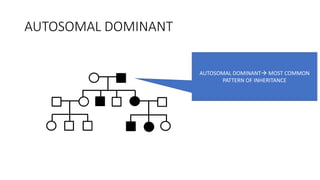
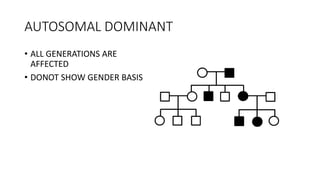
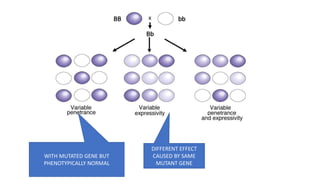
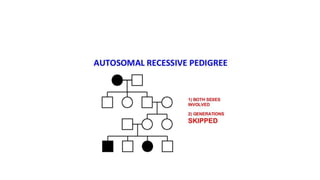
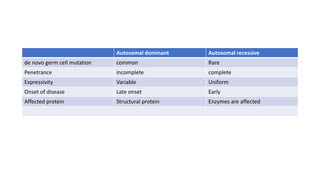
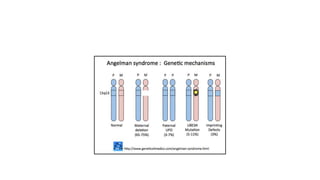
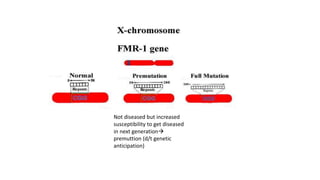
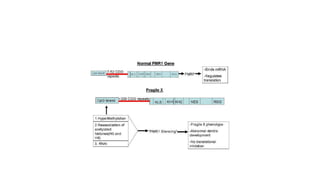
- Different patterns of inheritance for genetic disorders like autosomal dominant, autosomal recessive, and X-linked inheritance.
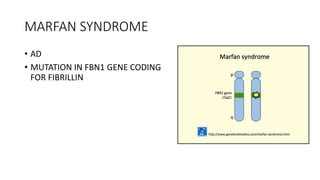
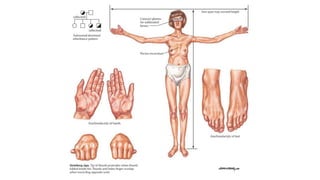
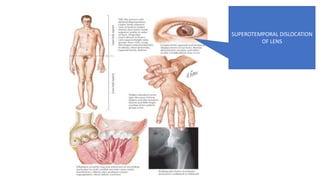
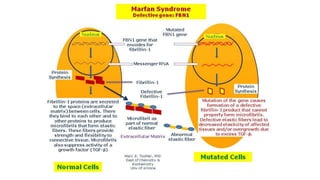
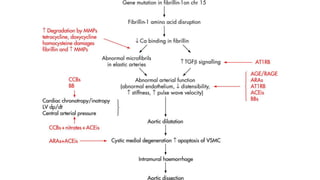
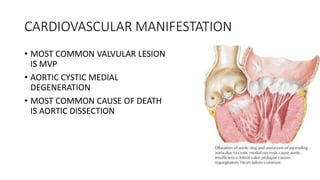
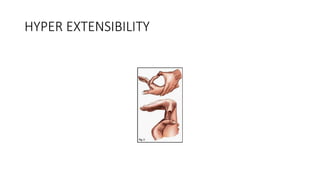
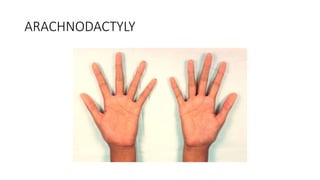
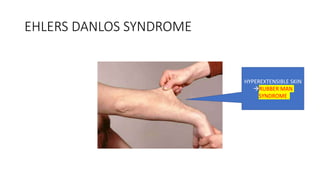
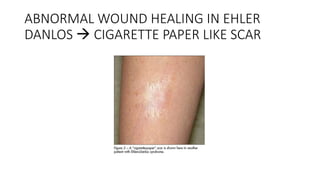
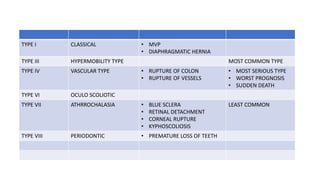
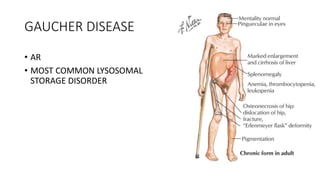
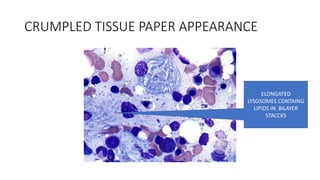
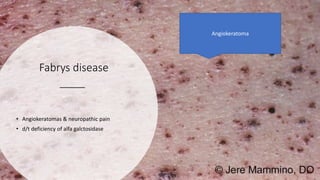
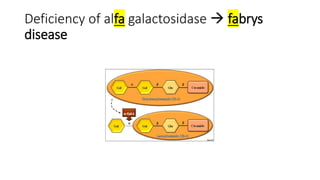
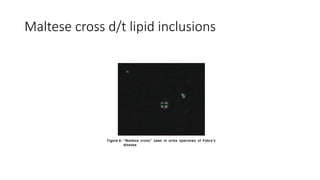
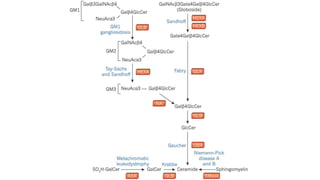
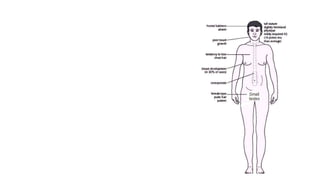
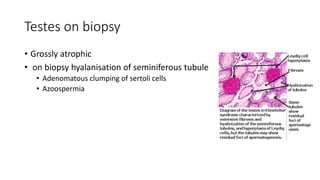
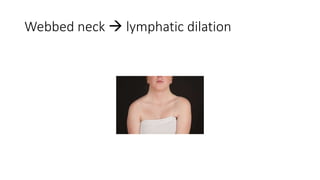
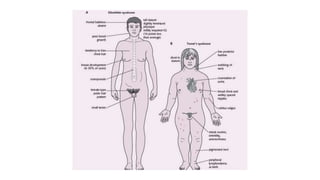
- Examples of genetic disorders and their inheritance patterns including Marfan syndrome, Ehlers-Danlos syndrome, and Fabry disease