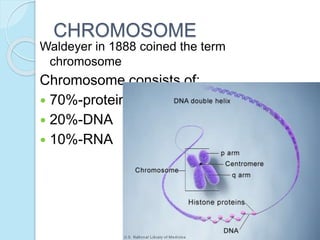
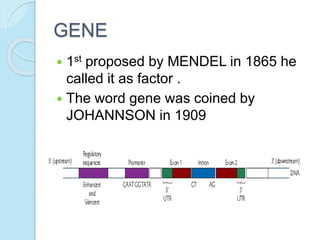
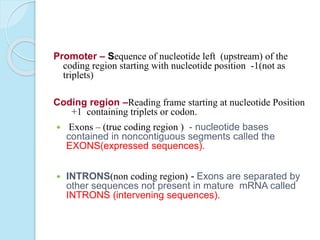
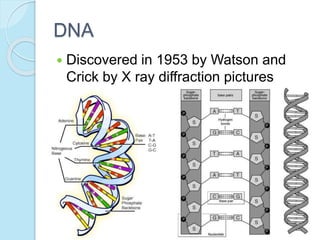
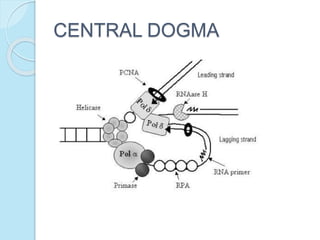
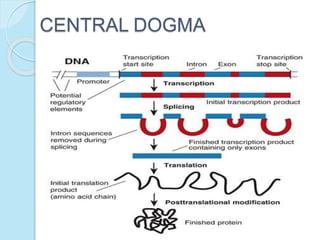
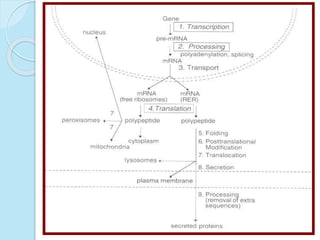
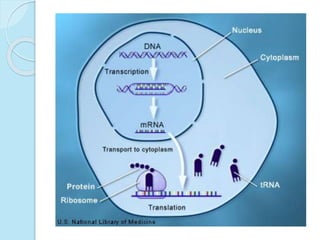
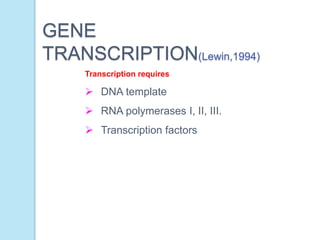
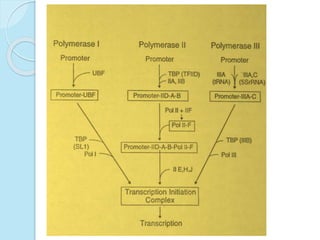
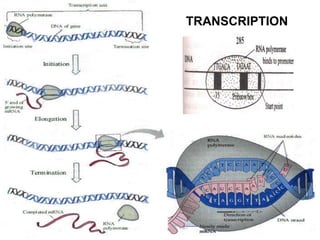
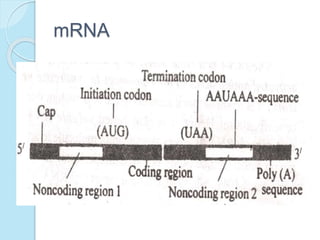
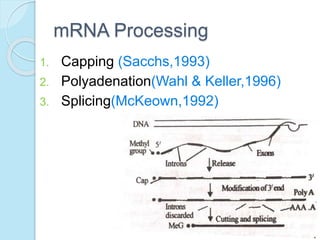
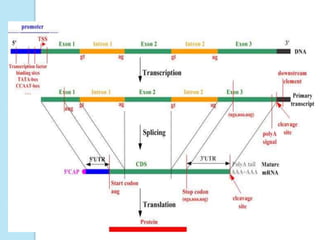
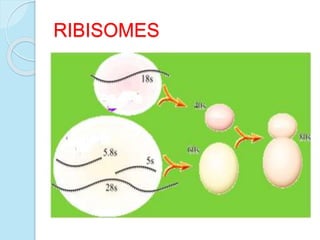
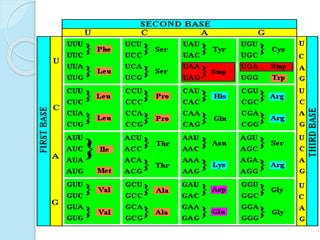
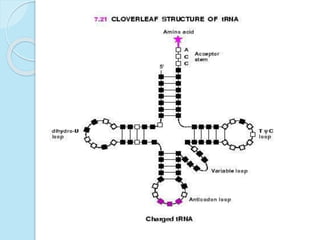
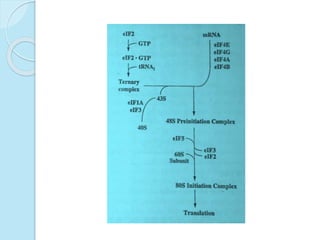
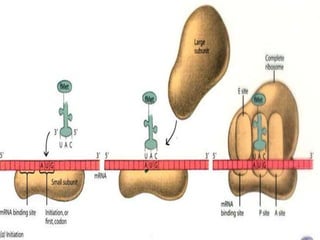
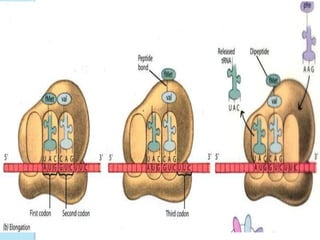
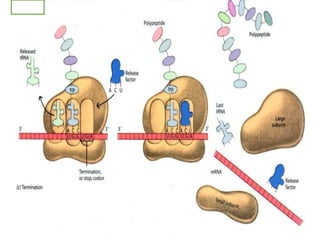
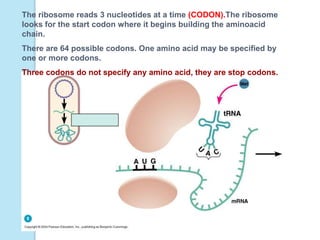
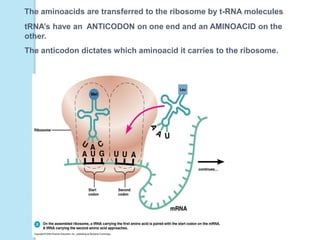
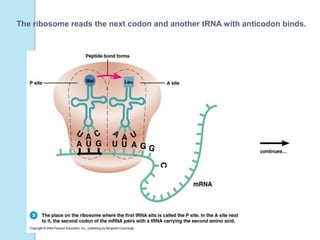
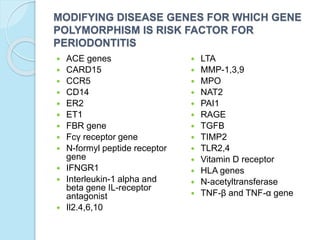
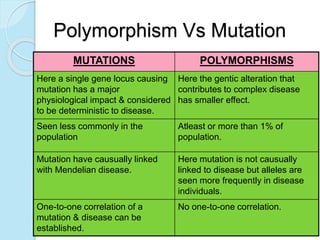
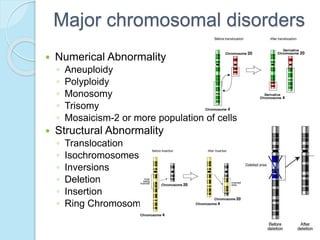
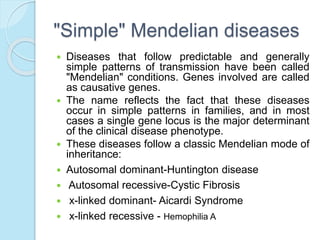
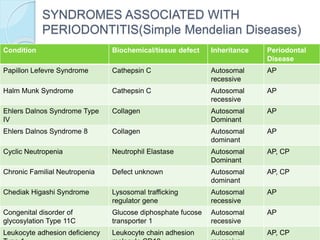
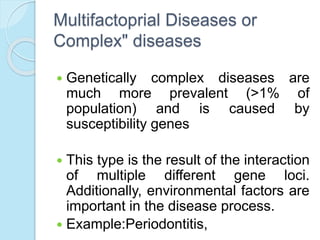
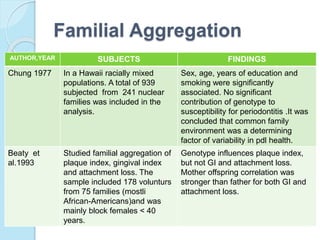
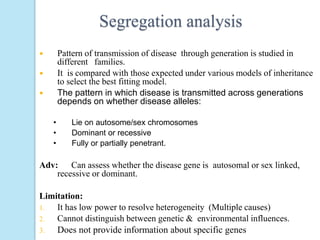
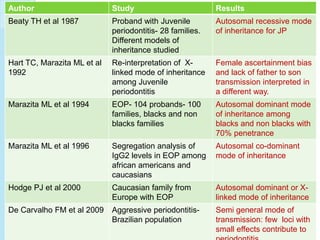
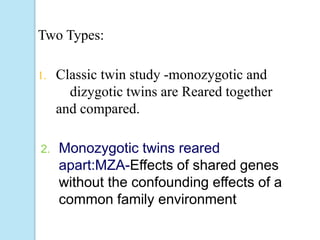
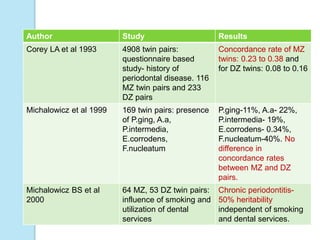
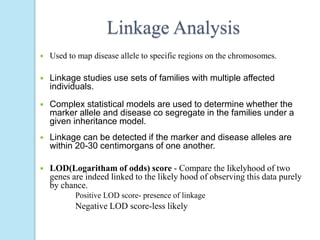
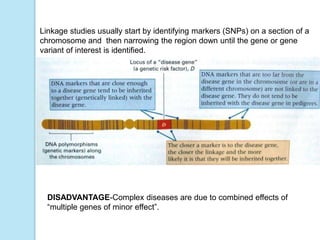
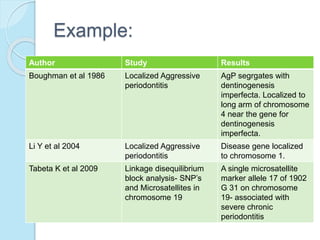
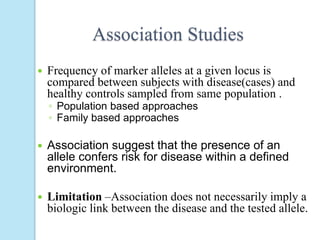
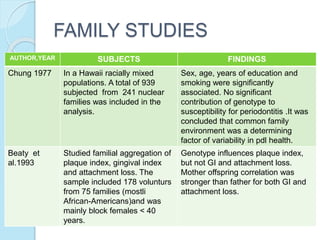
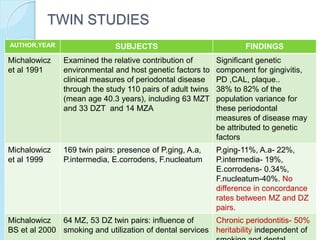
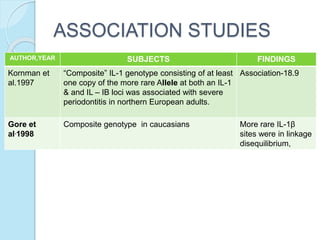
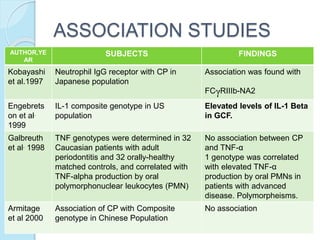
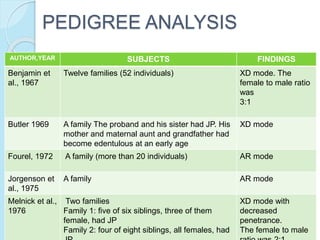
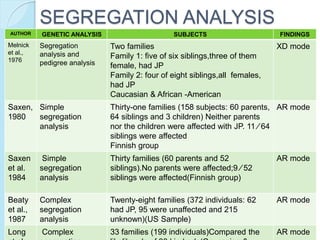
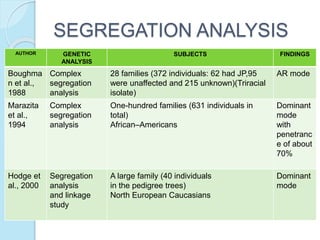
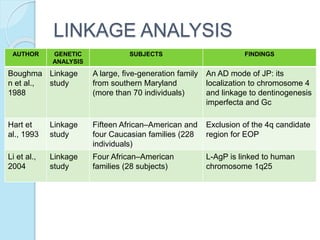
The document outlines a comprehensive study on genetics, focusing on heredity, gene structure, and gene expression, with specific emphasis on its relationship to periodontal diseases such as chronic and aggressive periodontitis. It details terminology relevant to genetics, various genetic mutations, polymorphisms, and the types of genetic disorders, while discussing their implications in understanding disease susceptibility and progression. Study designs for examining genetic factors include familial aggregation, segregation analysis, twin studies, and others, highlighting the complexity of genetics in relation to human health.