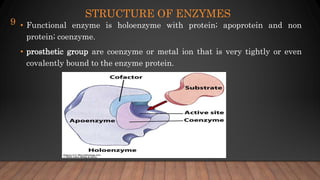
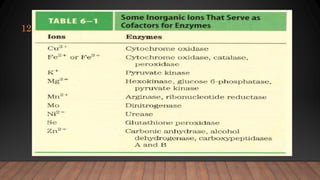
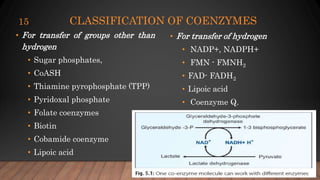
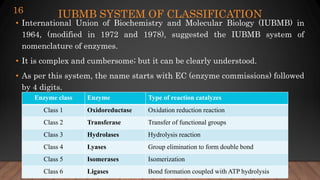
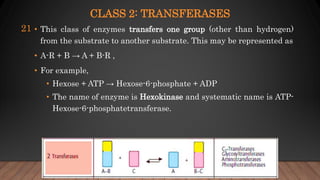
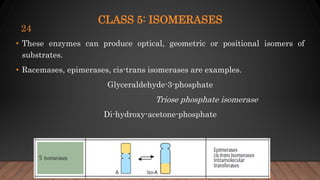
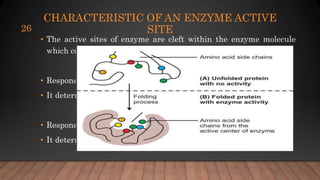
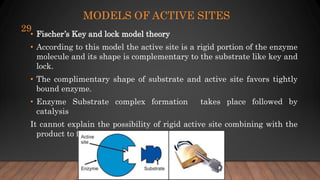
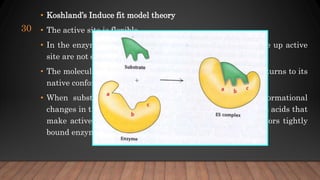
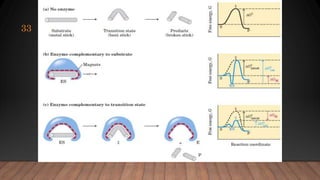
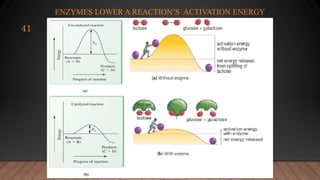
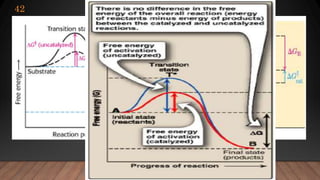
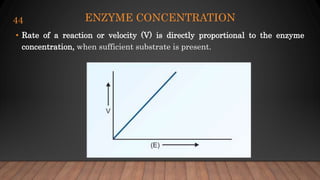
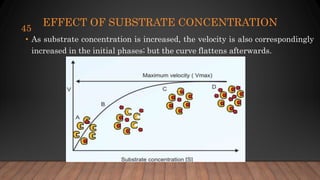
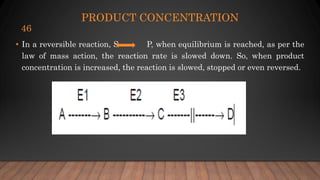
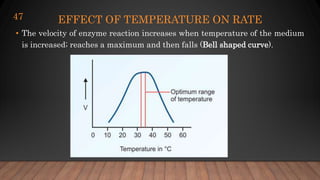
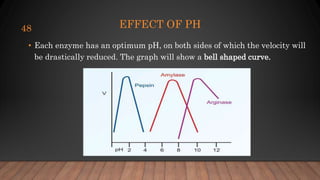
The document provides an overview of enzymes, their history, definitions, chemical nature, structure, classification, and mechanisms of action. Enzymes are biocatalysts essential for biochemical processes, classified into six main classes based on the type of reaction they catalyze. Key concepts include enzyme-substrate interactions and factors affecting enzyme activity, emphasizing their role in increasing reaction rates without being consumed.