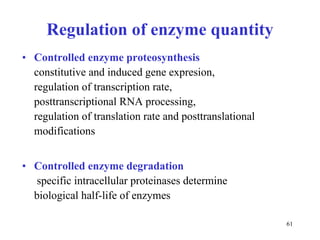
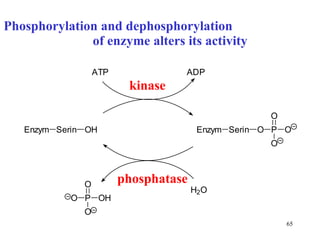
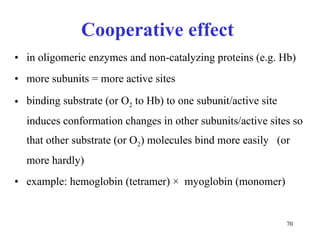
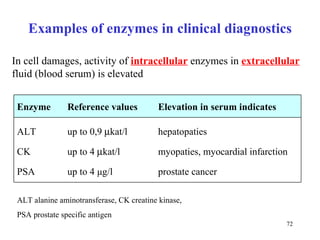
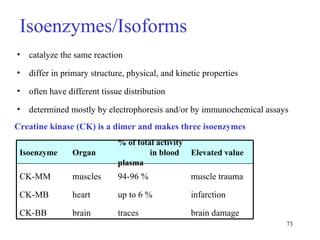
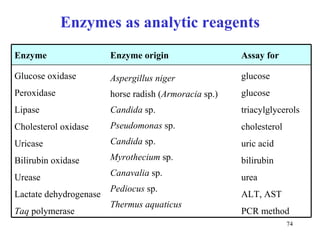
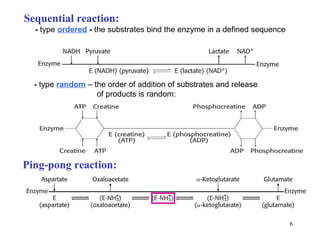
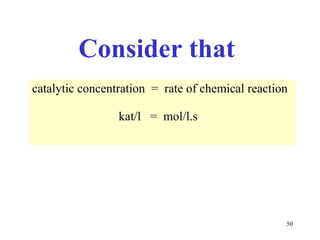
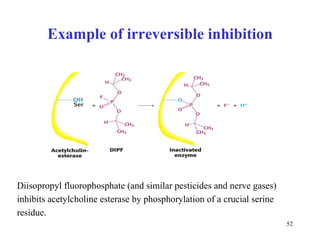
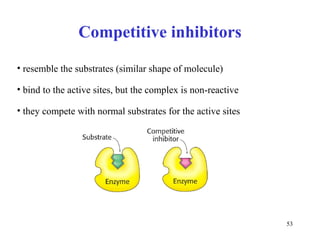
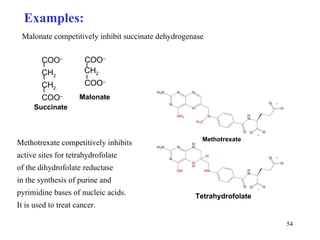
Enzymes catalyze biochemical reactions and are characterized by their catalytic activity. The document discusses several key aspects of enzyme mechanisms and kinetics, including:
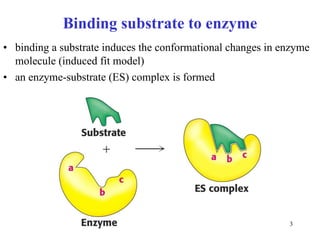
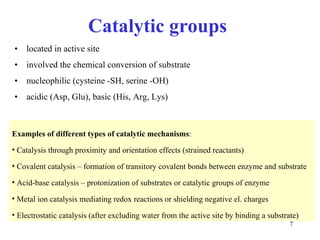
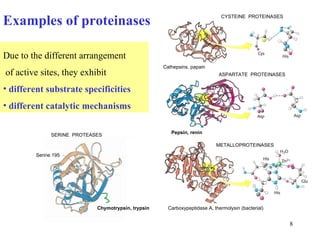
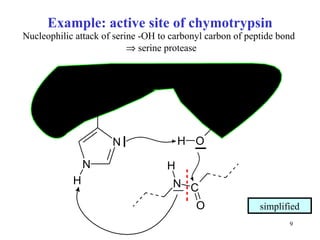
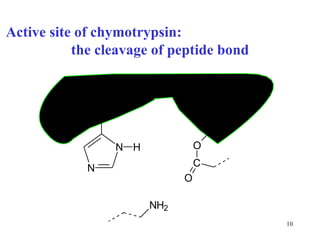
1) Enzymes have an active site that binds substrates and facilitates chemical reactions through proximity, orientation, and catalytic groups like metal ions or amino acid residues.
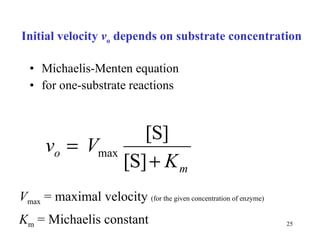
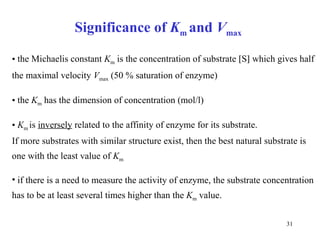
2) Michaelis-Menten kinetics describe the relationship between substrate concentration, reaction rate, and parameters like Vmax and Km.
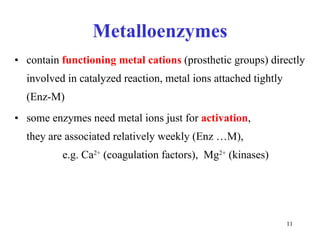
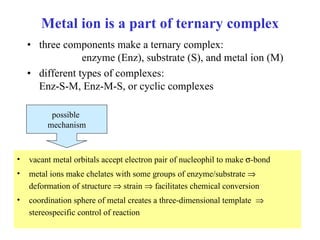
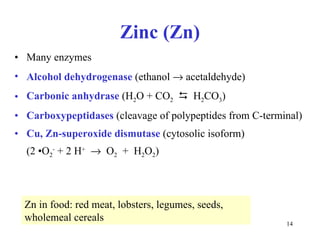
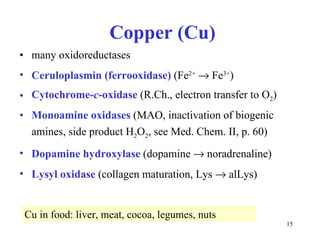
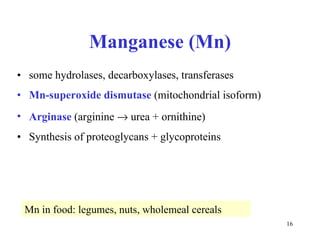
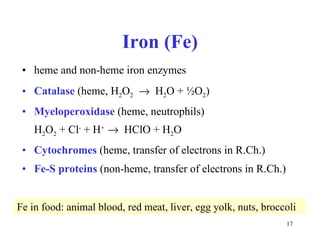
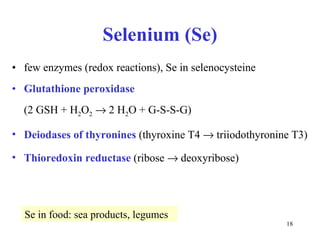
3) Common metalloenzymes contain catalytic metal ions like zinc, copper, iron and manganese that participate in redox reactions.


![Basic kinetic terms rea ction : S P (S = substr ate , P = produ c t) Defini tion of rea ction velocity (rate): this definiton is for average rate, instantaneous rate: d[S]/d t](https://image.slidesharecdn.com/enzymes-2-110919095503-phpapp01/85/Enzymes-2-19-320.jpg)
![Factors influencing rea ction rate concentra tion of substr ate [S] temperature t he presence of c ataly st or inhibitor In enzyme reactions c oncentration of enzyme [E] pH](https://image.slidesharecdn.com/enzymes-2-110919095503-phpapp01/85/Enzymes-2-20-320.jpg)
![Kinetic equation for rea ction S P v = k [S] = k [S] 1 reaction of 1. order k = rate c onstant](https://image.slidesharecdn.com/enzymes-2-110919095503-phpapp01/85/Enzymes-2-21-320.jpg)
![K inetic (progress) curve s for substrate and product during rea ction : concentra tion of substr ate decreases concentration of product increases reaction rate is determined from kinetic curves The instantaneous velocity v x at any particular time t x is g iven by the slope of the tangent to the curve at that time. [ P ] t [ S ] t (equilibrium)](https://image.slidesharecdn.com/enzymes-2-110919095503-phpapp01/85/Enzymes-2-22-320.jpg)
![Rea ction of 0. order Rea ction rate does not depend on the substrate c oncentra tion v = k [S] 0 = k × 1 = k = constant At great excess of substrate S in enzyme rections only in laboratory conditions](https://image.slidesharecdn.com/enzymes-2-110919095503-phpapp01/85/Enzymes-2-23-320.jpg)
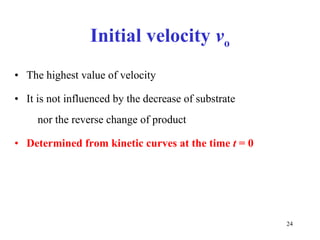
![If [S] << K m at low substrate concentration the reaction proceeds by the 1 st order kinetics ×](https://image.slidesharecdn.com/enzymes-2-110919095503-phpapp01/85/Enzymes-2-27-320.jpg)
![If [S] >> K m at high substrate concentration the reaction proceeds by the 0. order kinetics ×](https://image.slidesharecdn.com/enzymes-2-110919095503-phpapp01/85/Enzymes-2-28-320.jpg)
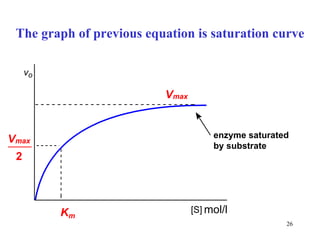
![Two parts of saturation curve compare with pages 21 and 23 [ S ] V 0 V max K m The zero-order kinetics The 1 st order kinetics](https://image.slidesharecdn.com/enzymes-2-110919095503-phpapp01/85/Enzymes-2-29-320.jpg)
![If [S] = K m](https://image.slidesharecdn.com/enzymes-2-110919095503-phpapp01/85/Enzymes-2-30-320.jpg)
![From the obtained progress curves, the values v o are estimated and plotted against the corresponding [S] . T he velocity v o rises linearly as substrate concentration increases, and then begins to level till it reaches a limit value at high substrate concentrations. How to get a saturation curve? E = const . A series of measurements of initial velocity is arranged at a constant enzyme concentration E and different substrate concentrations S (in the range of 2 - 3 orders). [ P ] t [ S 1 ] [ S 2 ] [ S 3 ] [ S 4 ] v 0 1 v 0 2 v 0 3 v 0 4](https://image.slidesharecdn.com/enzymes-2-110919095503-phpapp01/85/Enzymes-2-32-320.jpg)
![Distinguish Kinetic curve time record of one reaction [S] = f ( t ) or [P] = f ( t ) Saturation curve dependence obtained from many reactions ( see p revious page ) v o = f ( [S]) [S] …….. substr ate concentration [P] .......... product concentration f ………… function t ………… time v o ……….. initial velocity](https://image.slidesharecdn.com/enzymes-2-110919095503-phpapp01/85/Enzymes-2-33-320.jpg)
![V max and K m describe the kinetic properties of enzyme are hardly obtained from saturation curve easily obtained from linear double reciprocal plot Lineweaver-Burk: 1/ v o is plotted against 1/ [S]](https://image.slidesharecdn.com/enzymes-2-110919095503-phpapp01/85/Enzymes-2-34-320.jpg)
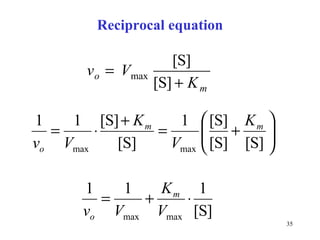
![Reciprocal form is the equation of a line ( y = a x + b) 1/ v o ................ dependent variable ( y ) 1/ [S] ............... independent variable ( x ) 1/ K m ........... .... 1/ V max ............. easily determined from the graph](https://image.slidesharecdn.com/enzymes-2-110919095503-phpapp01/85/Enzymes-2-36-320.jpg)
![Linear reciprocal plot: 1/ v o is the function of 1/ [S]](https://image.slidesharecdn.com/enzymes-2-110919095503-phpapp01/85/Enzymes-2-37-320.jpg)
![Initial velocity depends on enzym e concentration [E] v o [S] [E 1 ] [E 2 ] > [E 1 ] [E 3 ] > [E 2 ] K M does not change, V max increases with increased [E] saturated enzym e : v o = k [E] t [E] t is tot al c oncentra tion of enzym e](https://image.slidesharecdn.com/enzymes-2-110919095503-phpapp01/85/Enzymes-2-38-320.jpg)

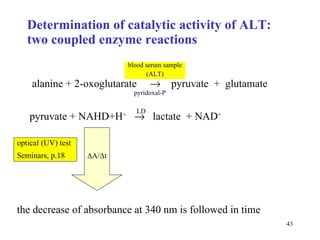
![Determination of catalytic activity in vitro optimal conditions: temperature, pH the presence of all necessary cofactors the absence of all (known) inhibitors excess of substrate 0. order kinetics: [S] >> K m saturated enzyme, reaction rate is constant and close to V max [S] or [P] is followed during time](https://image.slidesharecdn.com/enzymes-2-110919095503-phpapp01/85/Enzymes-2-44-320.jpg)
![Two methods for catalytic concentration less exact exact Evaluation of method average velocity initial velocity v o What is determined no yes Kinetic curve needed after some time (e.g. 10 min) the reaction is stopped by enzyme inactivation continually ( e.g. in 10 s econds ) How [P] [S] or [P] What is measured Constant-time method Kinetic method Feature](https://image.slidesharecdn.com/enzymes-2-110919095503-phpapp01/85/Enzymes-2-45-320.jpg)
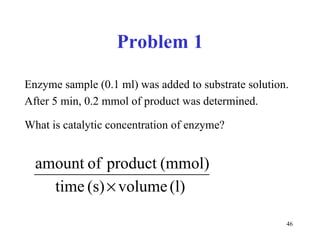

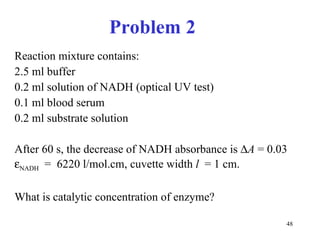
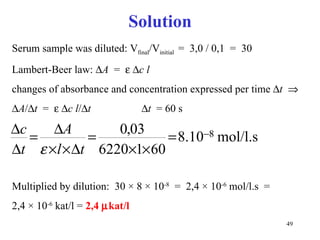

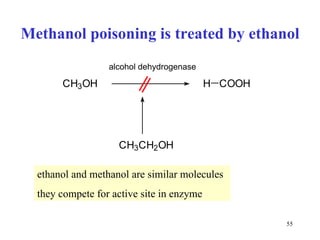
![Competitive inhibitors increase K m without any change in V max The V max can be reached even in the presence of inhibitor, but at much higher concentrations of [S] that have to overcome the competing inhibitor concentration. K m [S] v 0 V max K m 1 / v 0 1 / [S] 1 / V max - 1 / K m No inhibitor Competitive inhibitor](https://image.slidesharecdn.com/enzymes-2-110919095503-phpapp01/85/Enzymes-2-56-320.jpg)
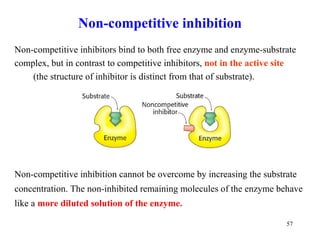
![Non - competitive inhibitors decrease V max without any change in K m 1/v o 1 / [S] 1 / V max - 1 / K m Non - competitive inhibitor No inhibitor K m [S] v 0 V max V max Non - competitive inhibitor](https://image.slidesharecdn.com/enzymes-2-110919095503-phpapp01/85/Enzymes-2-58-320.jpg)