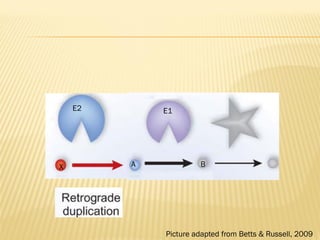
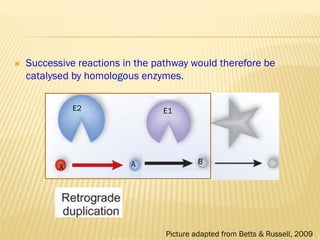
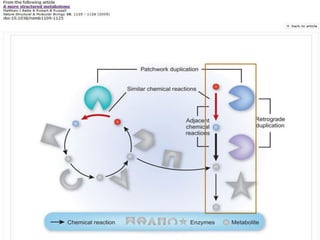
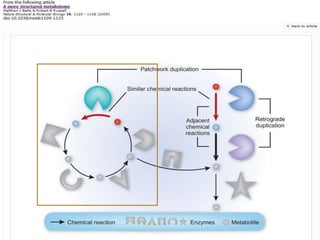
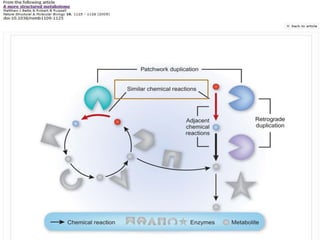
This document discusses two main theories for how enzyme systems evolved: retrograde evolution and patchwork evolution. Retrograde evolution proposes that pathways evolve backwards, with newer reactions preceding existing ones. Patchwork evolution suggests that enzymes initially had broad specificities and specialized over time through gene duplication. While some evidence supports retrograde evolution, more data favors patchwork evolution. However, the CS2 hydrolase enzyme appears to be an exception, as its structure and function indicate it did not evolve but was purposefully designed. In conclusion, the theories of enzyme evolution are still being refined as exceptions are discovered.













![͞EŶzyŵes ĐouldŶ’t eǀolǀe͟
*The investigators compared the three-dimensional structures of
similarly shaped enzymes that are found in different species of
bacteria:
One enzyme splits water and combines the resulting hydrogen
atoms with sulfur . (CS2 hydrolase]
Another class of enzymes that also splits water molecules, but then
combines the hydrogen with a carbon-based molecule.
*The core structure of the CS2 hydrolase like that of similar enzymes,
is critical. The scientists wrote in Nature, "Any change in this area
of the protein [enzyme] adversely affected protein activity."
*The researchers also found that CS2 hydrolase is distinct from
enzymes with an otherwise identical core because it has an
additional long, narrow tunnel through which only CS2 can pass.
The tunnel "functions as a specificity filter," ensuring that no
similar molecule such as carbon dioxide enters.](https://image.slidesharecdn.com/enzymesystemevolution-141014052946-conversion-gate01/85/Enzyme-system-evolution-22-320.jpg)